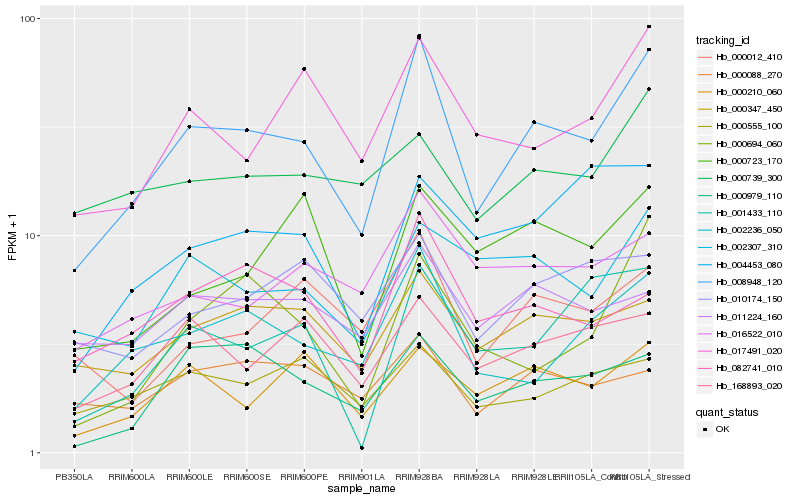
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008948_120 |
0.0 |
- |
- |
PREDICTED: heme oxygenase 1, chloroplastic-like [Jatropha curcas] |
| 2 |
Hb_002236_050 |
0.1357494214 |
- |
- |
- |
| 3 |
Hb_017491_020 |
0.1520586168 |
- |
- |
hypothetical protein B456_011G056700 [Gossypium raimondii] |
| 4 |
Hb_000347_450 |
0.1561346989 |
- |
- |
PREDICTED: tRNA(His) guanylyltransferase 1-like isoform X2 [Jatropha curcas] |
| 5 |
Hb_002307_310 |
0.1605501337 |
- |
- |
PREDICTED: outer envelope pore protein 37, chloroplastic [Jatropha curcas] |
| 6 |
Hb_168893_020 |
0.1611294451 |
- |
- |
PREDICTED: probable protein S-acyltransferase 15 [Jatropha curcas] |
| 7 |
Hb_000210_060 |
0.1628711995 |
transcription factor |
TF Family: E2F-DP |
hypothetical protein JCGZ_08780 [Jatropha curcas] |
| 8 |
Hb_000012_410 |
0.1648097472 |
- |
- |
PREDICTED: histone deacetylase 2 isoform X1 [Jatropha curcas] |
| 9 |
Hb_016522_010 |
0.1687963311 |
- |
- |
PREDICTED: short-chain dehydrogenase/reductase 2b-like [Jatropha curcas] |
| 10 |
Hb_082741_010 |
0.1689819066 |
- |
- |
PREDICTED: uncharacterized protein LOC105649930 [Jatropha curcas] |
| 11 |
Hb_000979_110 |
0.1704927133 |
- |
- |
PREDICTED: GPI mannosyltransferase 2 [Jatropha curcas] |
| 12 |
Hb_010174_150 |
0.1714182384 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 13 |
Hb_004453_080 |
0.1728694519 |
- |
- |
PREDICTED: acyl-protein thioesterase 2-like [Populus euphratica] |
| 14 |
Hb_011224_160 |
0.173342455 |
transcription factor, rubber biosynthesis |
TF Family: HSF, Gene Name: Farnesyl diphosphate synthase |
PREDICTED: heat stress transcription factor A-5 [Jatropha curcas] |
| 15 |
Hb_000555_100 |
0.1739457748 |
- |
- |
hypothetical protein JCGZ_14689 [Jatropha curcas] |
| 16 |
Hb_000694_060 |
0.1752683946 |
- |
- |
Rotenone-insensitive NADH-ubiquinone oxidoreductase, mitochondrial precursor, putative [Ricinus communis] |
| 17 |
Hb_000088_270 |
0.1762117044 |
- |
- |
PREDICTED: uncharacterized protein LOC105637003 [Jatropha curcas] |
| 18 |
Hb_001433_110 |
0.1793741556 |
- |
- |
PREDICTED: uncharacterized protein LOC105647469 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000723_170 |
0.1803957577 |
- |
- |
PREDICTED: OTU domain-containing protein DDB_G0284757 [Jatropha curcas] |
| 20 |
Hb_000739_300 |
0.1806398244 |
- |
- |
PREDICTED: uncharacterized protein LOC105643092 [Jatropha curcas] |