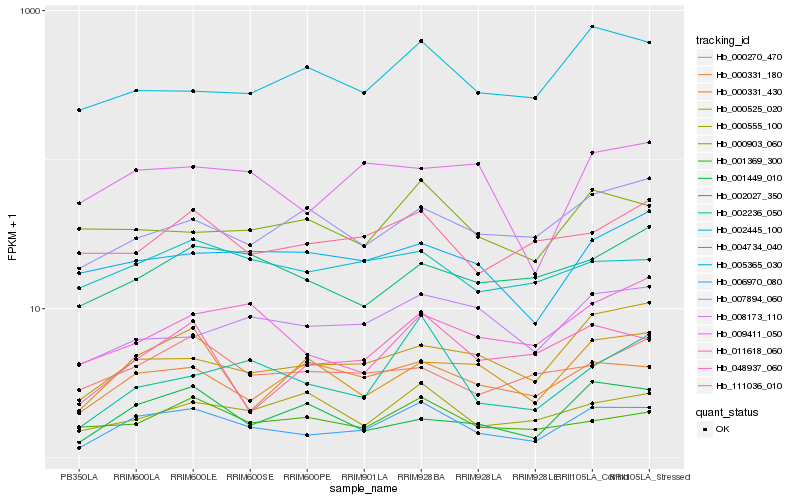
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006970_080 |
0.0 |
- |
- |
PREDICTED: putative lysine-specific demethylase JMJD5 [Jatropha curcas] |
| 2 |
Hb_000331_180 |
0.155924971 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105640220 [Jatropha curcas] |
| 3 |
Hb_048937_060 |
0.1580781971 |
- |
- |
PREDICTED: probable rhamnogalacturonate lyase B isoform X1 [Jatropha curcas] |
| 4 |
Hb_000525_020 |
0.1604458797 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 5 |
Hb_002236_050 |
0.1637501627 |
- |
- |
- |
| 6 |
Hb_001369_300 |
0.1640978384 |
- |
- |
PREDICTED: structural maintenance of chromosomes protein 2-1-like [Jatropha curcas] |
| 7 |
Hb_004734_040 |
0.1668503594 |
- |
- |
eukaryotic translation initiation factor 5A isoform I [Hevea brasiliensis] |
| 8 |
Hb_000331_430 |
0.1677035909 |
- |
- |
PREDICTED: uncharacterized protein LOC105640242 [Jatropha curcas] |
| 9 |
Hb_009411_050 |
0.1681441055 |
- |
- |
PREDICTED: protein EARLY RESPONSIVE TO DEHYDRATION 15 [Jatropha curcas] |
| 10 |
Hb_008173_110 |
0.1686626464 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 11 |
Hb_007894_060 |
0.1694058293 |
- |
- |
PREDICTED: ubiquitin thioesterase OTU1 [Jatropha curcas] |
| 12 |
Hb_002445_100 |
0.1695529287 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 5 isoform X2 [Jatropha curcas] |
| 13 |
Hb_002027_350 |
0.1697456813 |
- |
- |
PREDICTED: U1 small nuclear ribonucleoprotein C-like [Jatropha curcas] |
| 14 |
Hb_000903_060 |
0.1707820606 |
- |
- |
PREDICTED: ras-related protein RABA5e [Jatropha curcas] |
| 15 |
Hb_005365_030 |
0.1709199192 |
- |
- |
hypothetical protein POPTR_0002s24010g [Populus trichocarpa] |
| 16 |
Hb_011618_060 |
0.1711676921 |
- |
- |
PREDICTED: DNA-repair protein XRCC1 [Jatropha curcas] |
| 17 |
Hb_001449_010 |
0.1716585651 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000555_100 |
0.1718253205 |
- |
- |
hypothetical protein JCGZ_14689 [Jatropha curcas] |
| 19 |
Hb_111036_010 |
0.1720437548 |
- |
- |
Eukaryotic translation initiation factor 6-2 [Glycine soja] |
| 20 |
Hb_000270_470 |
0.1722087907 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: pentatricopeptide repeat-containing protein At1g74900, mitochondrial [Jatropha curcas] |