| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
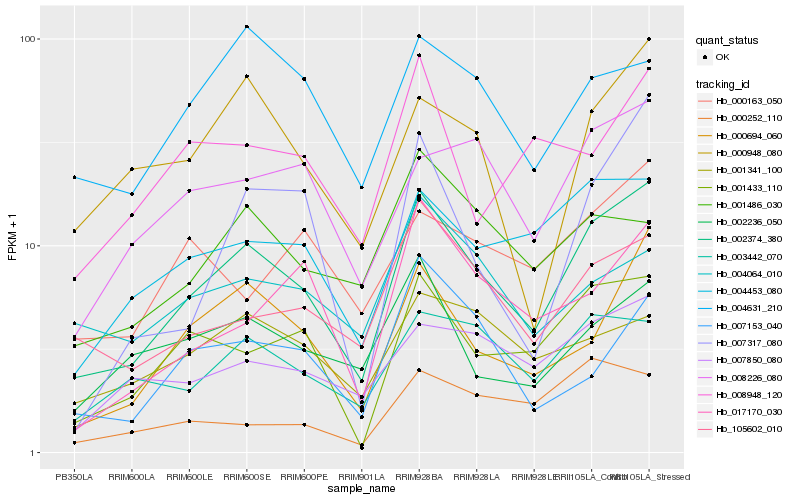
Hb_002374_380 |
0.0 |
- |
- |
Acyl-CoA N-acyltransferases (NAT) superfamily protein isoform 1 [Theobroma cacao] |
| 2 |
Hb_000694_060 |
0.14672013 |
- |
- |
Rotenone-insensitive NADH-ubiquinone oxidoreductase, mitochondrial precursor, putative [Ricinus communis] |
| 3 |
Hb_001433_110 |
0.1477237999 |
- |
- |
PREDICTED: uncharacterized protein LOC105647469 isoform X1 [Jatropha curcas] |
| 4 |
Hb_004453_080 |
0.1524655362 |
- |
- |
PREDICTED: acyl-protein thioesterase 2-like [Populus euphratica] |
| 5 |
Hb_105602_010 |
0.1564778678 |
- |
- |
Ketose-bisphosphate aldolase class-II family protein isoform 5 [Theobroma cacao] |
| 6 |
Hb_003442_070 |
0.1619614186 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase SD1-8 [Jatropha curcas] |
| 7 |
Hb_007850_080 |
0.1635335134 |
- |
- |
PREDICTED: auxilin-like protein 1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_008226_080 |
0.1659074259 |
- |
- |
Protein LRP16, putative [Ricinus communis] |
| 9 |
Hb_002236_050 |
0.1687431421 |
- |
- |
- |
| 10 |
Hb_000948_080 |
0.1698949674 |
- |
- |
PREDICTED: uncharacterized protein LOC105634692 [Jatropha curcas] |
| 11 |
Hb_017170_030 |
0.1806096855 |
- |
- |
PREDICTED: acyl-coenzyme A oxidase 3, peroxisomal-like [Jatropha curcas] |
| 12 |
Hb_004631_210 |
0.1843198491 |
- |
- |
hypothetical protein POPTR_0007s05070g [Populus trichocarpa] |
| 13 |
Hb_001486_030 |
0.1852198424 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001341_100 |
0.1852297673 |
- |
- |
hypothetical protein JCGZ_05568 [Jatropha curcas] |
| 15 |
Hb_008948_120 |
0.18561584 |
- |
- |
PREDICTED: heme oxygenase 1, chloroplastic-like [Jatropha curcas] |
| 16 |
Hb_007153_040 |
0.1862541412 |
- |
- |
PREDICTED: uncharacterized protein LOC105633515 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000163_050 |
0.186314557 |
- |
- |
SPLICEOSOMAL protein U1A [Populus trichocarpa] |
| 18 |
Hb_007317_080 |
0.1873567911 |
- |
- |
PREDICTED: ferredoxin, root R-B2 [Jatropha curcas] |
| 19 |
Hb_000252_110 |
0.1884778616 |
- |
- |
- |
| 20 |
Hb_004064_010 |
0.1895356228 |
- |
- |
PREDICTED: uncharacterized protein LOC105133340 isoform X1 [Populus euphratica] |