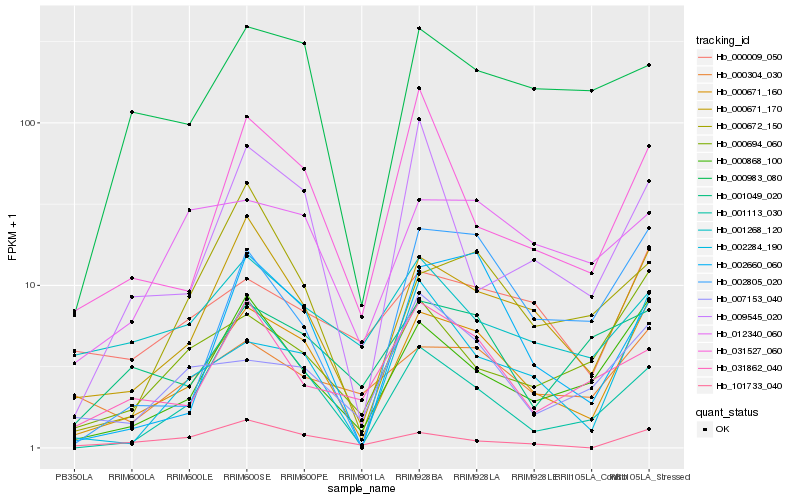
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000671_160 |
0.0 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000671_170 |
0.1388915857 |
- |
- |
kinase, putative [Ricinus communis] |
| 3 |
Hb_000868_100 |
0.1622911781 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RNF144A [Jatropha curcas] |
| 4 |
Hb_002284_190 |
0.1912017231 |
- |
- |
PREDICTED: glutamate receptor 3.3 [Jatropha curcas] |
| 5 |
Hb_000694_060 |
0.1995956179 |
- |
- |
Rotenone-insensitive NADH-ubiquinone oxidoreductase, mitochondrial precursor, putative [Ricinus communis] |
| 6 |
Hb_002660_060 |
0.203963538 |
- |
- |
PREDICTED: uncharacterized protein LOC105642215 [Jatropha curcas] |
| 7 |
Hb_002805_020 |
0.2043891618 |
transcription factor |
TF Family: SRS |
transcription factor, putative [Ricinus communis] |
| 8 |
Hb_031527_060 |
0.2159725045 |
- |
- |
glutamate decarboxylase, putative [Ricinus communis] |
| 9 |
Hb_000009_050 |
0.2165543482 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein indeterminate-domain 2-like [Jatropha curcas] |
| 10 |
Hb_007153_040 |
0.2166839022 |
- |
- |
PREDICTED: uncharacterized protein LOC105633515 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000304_030 |
0.2168877286 |
- |
- |
PREDICTED: seed maturation protein PM36 [Jatropha curcas] |
| 12 |
Hb_000983_080 |
0.2174910108 |
transcription factor |
TF Family: bZIP |
PREDICTED: ocs element-binding factor 1-like [Jatropha curcas] |
| 13 |
Hb_101733_040 |
0.2177296892 |
- |
- |
cytochrome P450 CYP71B63v2 [Populus trichocarpa] |
| 14 |
Hb_012340_060 |
0.2201184417 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001049_020 |
0.2225042236 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_031862_040 |
0.2332007066 |
- |
- |
purine transporter, putative [Ricinus communis] |
| 17 |
Hb_001113_030 |
0.2350746337 |
- |
- |
PREDICTED: transmembrane protein 45A-like [Jatropha curcas] |
| 18 |
Hb_001268_120 |
0.2386880682 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000672_150 |
0.2406935949 |
- |
- |
Patatin T5 precursor, putative [Ricinus communis] |
| 20 |
Hb_009545_020 |
0.2409565834 |
- |
- |
PREDICTED: uncharacterized protein LOC105632008 [Jatropha curcas] |