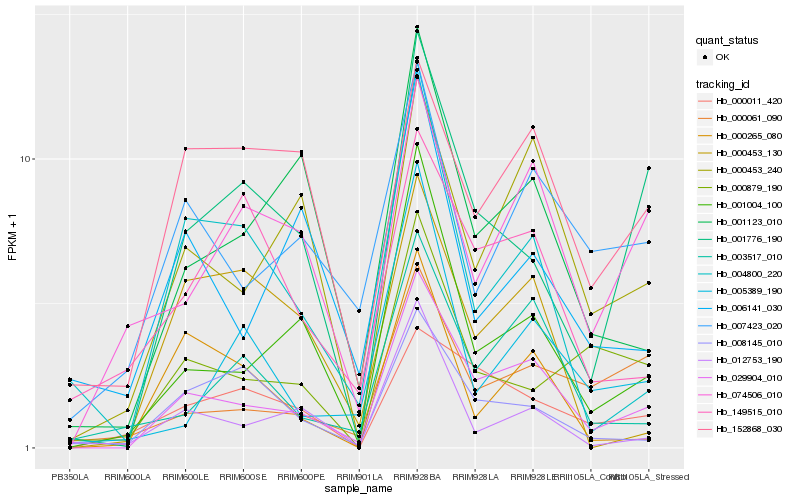
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_029904_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105635075 [Jatropha curcas] |
| 2 |
Hb_001004_100 |
0.1655301211 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 3 |
Hb_000453_240 |
0.1862769574 |
rubber biosynthesis |
Gene Name: 4-(cytidine 5'-diphospho)-2-C-methyl-D-erythritol kinase |
4-(cytidine 5'-diphospho)-2-C-methyl-D-erythritol kinase [Hevea brasiliensis] |
| 4 |
Hb_001123_010 |
0.1961895547 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_003517_010 |
0.2031078161 |
- |
- |
PREDICTED: ABC transporter C family member 3-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_001776_190 |
0.2062990095 |
- |
- |
PREDICTED: protein kinase 2A, chloroplastic-like [Jatropha curcas] |
| 7 |
Hb_000061_090 |
0.2138872091 |
- |
- |
hypothetical protein B456_003G0981001, partial [Gossypium raimondii] |
| 8 |
Hb_007423_020 |
0.2160214789 |
- |
- |
2-nitropropane dioxygenase precursor, putative [Ricinus communis] |
| 9 |
Hb_004800_220 |
0.2178998597 |
- |
- |
PREDICTED: random slug protein 5 [Jatropha curcas] |
| 10 |
Hb_000011_420 |
0.2202715282 |
- |
- |
PREDICTED: GDSL esterase/lipase At1g29660-like [Citrus sinensis] |
| 11 |
Hb_074506_010 |
0.2239441503 |
- |
- |
JHL25H03.10 [Jatropha curcas] |
| 12 |
Hb_012753_190 |
0.226117784 |
- |
- |
PREDICTED: protein HAPLESS 2 [Populus euphratica] |
| 13 |
Hb_000879_190 |
0.2348207586 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_008145_010 |
0.2351021932 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 15 |
Hb_152868_030 |
0.2358320314 |
- |
- |
abhydrolase domain containing, putative [Ricinus communis] |
| 16 |
Hb_000265_080 |
0.2401852089 |
- |
- |
Vacuolar cation/proton exchanger, putative [Ricinus communis] |
| 17 |
Hb_005389_190 |
0.2408069914 |
- |
- |
glutamate receptor 2 plant, putative [Ricinus communis] |
| 18 |
Hb_006141_030 |
0.2447734187 |
- |
- |
- |
| 19 |
Hb_149515_010 |
0.2461524243 |
- |
- |
multidrug resistance-associated protein 2, 6 (mrp2, 6), abc-transoprter, putative [Ricinus communis] |
| 20 |
Hb_000453_130 |
0.2461960064 |
- |
- |
PREDICTED: protein kinase 2A, chloroplastic-like [Jatropha curcas] |