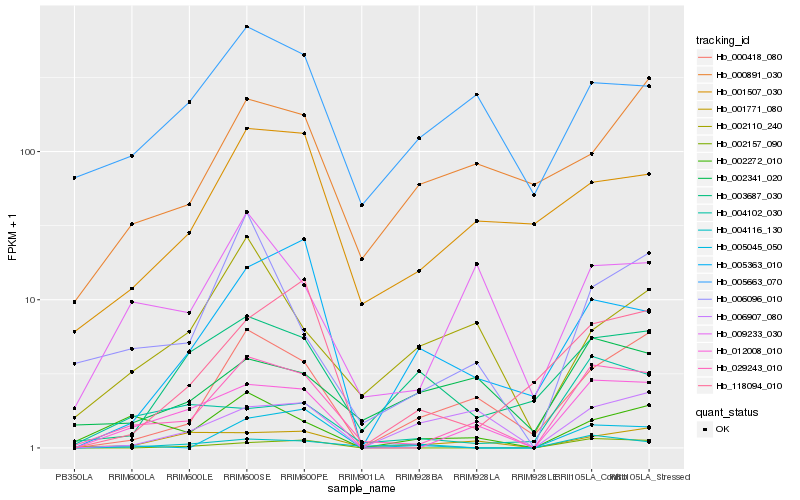
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_029243_010 |
0.0 |
- |
- |
PREDICTED: protein LURP-one-related 15-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_012008_010 |
0.1433934261 |
- |
- |
- |
| 3 |
Hb_000418_080 |
0.2173226238 |
- |
- |
PREDICTED: uncharacterized protein LOC105124652 [Populus euphratica] |
| 4 |
Hb_009233_030 |
0.2293958895 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 5 |
Hb_001507_030 |
0.2465124068 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 2 [Jatropha curcas] |
| 6 |
Hb_001771_080 |
0.2481252308 |
- |
- |
PREDICTED: UDP-D-apiose/UDP-D-xylose synthase 2 [Amborella trichopoda] |
| 7 |
Hb_002272_010 |
0.2486768971 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_006096_010 |
0.2507773504 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_005663_070 |
0.2552476501 |
- |
- |
dehydrin protein [Manihot esculenta] |
| 10 |
Hb_002157_090 |
0.2586683426 |
- |
- |
hypothetical protein VITISV_041741 [Vitis vinifera] |
| 11 |
Hb_002341_020 |
0.2616796255 |
- |
- |
PREDICTED: pyruvate decarboxylase 2 [Populus euphratica] |
| 12 |
Hb_004116_130 |
0.2753733362 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 13 |
Hb_003687_030 |
0.275796003 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 14 |
Hb_005045_050 |
0.2813802786 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein 3 [Jatropha curcas] |
| 15 |
Hb_118094_010 |
0.2822895182 |
- |
- |
hypothetical protein POPTR_0011s06595g [Populus trichocarpa] |
| 16 |
Hb_004102_030 |
0.2834040091 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At5g63020 [Jatropha curcas] |
| 17 |
Hb_006907_080 |
0.2843683584 |
- |
- |
PREDICTED: RPM1-interacting protein 4-like [Jatropha curcas] |
| 18 |
Hb_000891_030 |
0.2851758595 |
- |
- |
PREDICTED: uncharacterized protein At4g22758 [Jatropha curcas] |
| 19 |
Hb_005363_010 |
0.2854889936 |
- |
- |
hypothetical protein JCGZ_13835 [Jatropha curcas] |
| 20 |
Hb_002110_240 |
0.2866286991 |
- |
- |
clathrin assembly protein, putative [Ricinus communis] |