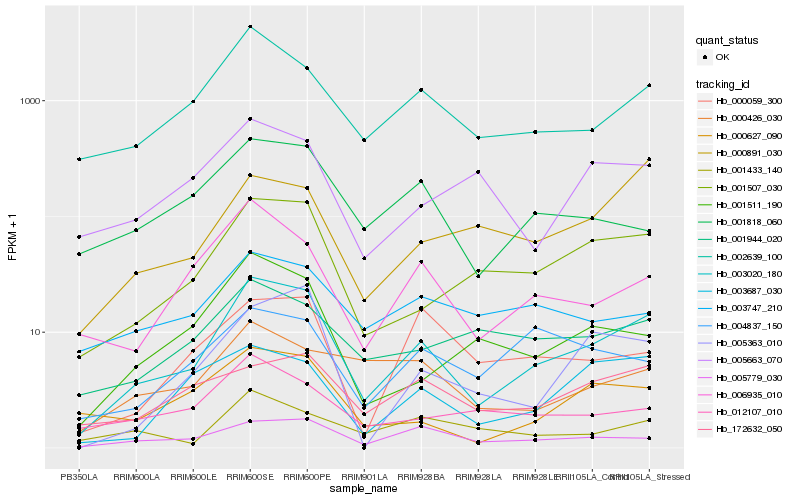
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003020_180 |
0.0 |
- |
- |
PREDICTED: probable protein phosphatase 2C 24 [Jatropha curcas] |
| 2 |
Hb_001507_030 |
0.1687492451 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 2 [Jatropha curcas] |
| 3 |
Hb_002639_100 |
0.1748297136 |
- |
- |
dehydrin [Hevea brasiliensis] |
| 4 |
Hb_006935_010 |
0.191404911 |
- |
- |
PREDICTED: inositol transporter 1 isoform X1 [Gossypium raimondii] |
| 5 |
Hb_005779_030 |
0.1921675675 |
- |
- |
PREDICTED: nuclear pore complex protein NUP62 [Jatropha curcas] |
| 6 |
Hb_000627_090 |
0.2039135568 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_003687_030 |
0.2063284788 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 8 |
Hb_001511_190 |
0.2075432822 |
- |
- |
PREDICTED: probable protein phosphatase 2C 25 [Jatropha curcas] |
| 9 |
Hb_172632_050 |
0.2127113623 |
- |
- |
PREDICTED: uncharacterized protein LOC105646134 [Jatropha curcas] |
| 10 |
Hb_005363_010 |
0.2140088677 |
- |
- |
hypothetical protein JCGZ_13835 [Jatropha curcas] |
| 11 |
Hb_001818_060 |
0.2142446743 |
- |
- |
PREDICTED: cysteine proteinase inhibitor 5-like [Jatropha curcas] |
| 12 |
Hb_001944_020 |
0.218402529 |
- |
- |
PREDICTED: uncharacterized protein LOC105636535 [Jatropha curcas] |
| 13 |
Hb_012107_010 |
0.2199872547 |
- |
- |
hypothetical protein JCGZ_19531 [Jatropha curcas] |
| 14 |
Hb_000891_030 |
0.2235924738 |
- |
- |
PREDICTED: uncharacterized protein At4g22758 [Jatropha curcas] |
| 15 |
Hb_001433_140 |
0.2273650376 |
- |
- |
PREDICTED: phospholipase D p1 [Jatropha curcas] |
| 16 |
Hb_000059_300 |
0.2278198566 |
- |
- |
caspase, putative [Ricinus communis] |
| 17 |
Hb_000426_030 |
0.2285888518 |
- |
- |
PREDICTED: protein STICHEL-like 3 isoform X2 [Jatropha curcas] |
| 18 |
Hb_003747_210 |
0.228597489 |
- |
- |
PREDICTED: uncharacterized protein LOC105630240 [Jatropha curcas] |
| 19 |
Hb_004837_150 |
0.2289775566 |
- |
- |
hypothetical protein POPTR_0001s26540g [Populus trichocarpa] |
| 20 |
Hb_005663_070 |
0.2304955139 |
- |
- |
dehydrin protein [Manihot esculenta] |