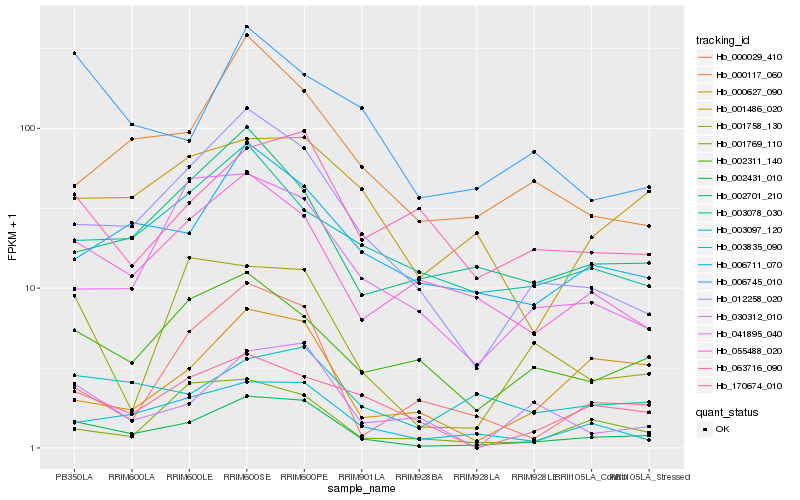
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002431_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104599557 isoform X2 [Nelumbo nucifera] |
| 2 |
Hb_012258_020 |
0.1667649075 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 3 |
Hb_003835_090 |
0.1718680917 |
transcription factor |
TF Family: AP2 |
PREDICTED: AP2-like ethylene-responsive transcription factor AIL6 [Jatropha curcas] |
| 4 |
Hb_006711_070 |
0.1945948103 |
- |
- |
hypothetical protein RCOM_1336320 [Ricinus communis] |
| 5 |
Hb_055488_020 |
0.1962602849 |
- |
- |
PREDICTED: uncharacterized protein LOC105641270 [Jatropha curcas] |
| 6 |
Hb_030312_010 |
0.198721689 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_170674_010 |
0.2006085775 |
- |
- |
hypothetical protein JCGZ_23765 [Jatropha curcas] |
| 8 |
Hb_002311_140 |
0.2049817431 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 15-like [Jatropha curcas] |
| 9 |
Hb_001486_020 |
0.2070904412 |
- |
- |
hypothetical protein JCGZ_02719 [Jatropha curcas] |
| 10 |
Hb_000117_060 |
0.208325703 |
- |
- |
PREDICTED: uncharacterized protein LOC103949199 [Pyrus x bretschneideri] |
| 11 |
Hb_002701_210 |
0.2085995062 |
- |
- |
Protein COBRA precursor, putative [Ricinus communis] |
| 12 |
Hb_063716_090 |
0.2087218193 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000627_090 |
0.2090319352 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001758_130 |
0.212199118 |
- |
- |
hypothetical protein JCGZ_25129 [Jatropha curcas] |
| 15 |
Hb_003097_120 |
0.2159208583 |
- |
- |
PREDICTED: cytokinin dehydrogenase 6 [Jatropha curcas] |
| 16 |
Hb_001769_110 |
0.2159276409 |
- |
- |
PREDICTED: probable metal-nicotianamine transporter YSL7 [Jatropha curcas] |
| 17 |
Hb_041895_040 |
0.2174341788 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_003078_030 |
0.2176521045 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase CHIP [Jatropha curcas] |
| 19 |
Hb_000029_410 |
0.2199279663 |
- |
- |
AT14A, putative [Ricinus communis] |
| 20 |
Hb_006745_010 |
0.2200559004 |
- |
- |
PREDICTED: uncharacterized protein LOC105640095 [Jatropha curcas] |