| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
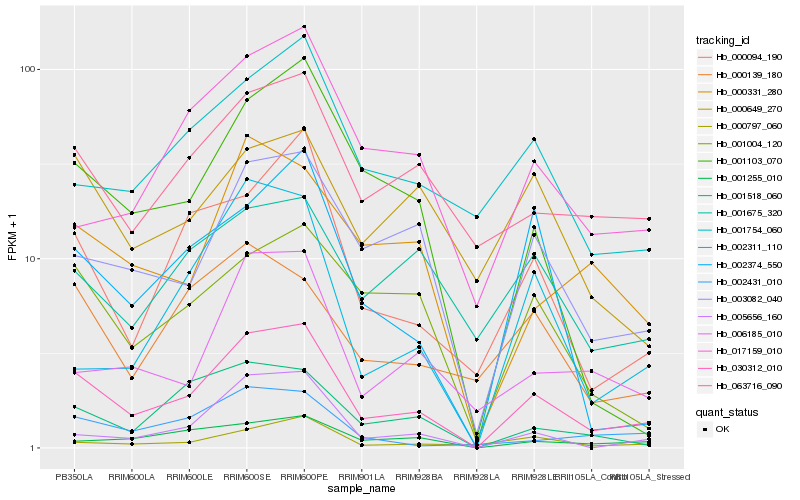
Hb_030312_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_003082_040 |
0.1610216552 |
- |
- |
PREDICTED: receptor-like protein kinase HSL1 [Phoenix dactylifera] |
| 3 |
Hb_000094_190 |
0.1762938275 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 7-like [Jatropha curcas] |
| 4 |
Hb_063716_090 |
0.1769838805 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001255_010 |
0.1855737242 |
- |
- |
PREDICTED: phytosulfokine receptor 1-like [Tarenaya hassleriana] |
| 6 |
Hb_000797_060 |
0.1883261199 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 7 |
Hb_001004_120 |
0.1886940206 |
- |
- |
PREDICTED: receptor-like protein kinase HERK 1 isoform X2 [Jatropha curcas] |
| 8 |
Hb_001518_060 |
0.1896264333 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 9 [Jatropha curcas] |
| 9 |
Hb_005656_160 |
0.1898675836 |
- |
- |
PREDICTED: uncharacterized protein LOC105637448 [Jatropha curcas] |
| 10 |
Hb_000331_280 |
0.1937019384 |
- |
- |
AMP dependent CoA ligase, putative [Ricinus communis] |
| 11 |
Hb_001754_060 |
0.1951179729 |
- |
- |
Xyloglucan 6-xylosyltransferase, putative [Ricinus communis] |
| 12 |
Hb_001103_070 |
0.1953483505 |
- |
- |
Patellin-3, putative [Ricinus communis] |
| 13 |
Hb_002431_010 |
0.198721689 |
- |
- |
PREDICTED: uncharacterized protein LOC104599557 isoform X2 [Nelumbo nucifera] |
| 14 |
Hb_002374_550 |
0.1989832904 |
- |
- |
PREDICTED: aspartic proteinase nepenthesin-1 [Jatropha curcas] |
| 15 |
Hb_006185_010 |
0.1999445236 |
- |
- |
hypothetical protein L484_013453 [Morus notabilis] |
| 16 |
Hb_000139_180 |
0.2015263774 |
- |
- |
PREDICTED: putative leucine-rich repeat receptor-like serine/threonine-protein kinase At2g14440 [Jatropha curcas] |
| 17 |
Hb_001675_320 |
0.2040560529 |
- |
- |
PREDICTED: uncharacterized protein LOC105635025 [Jatropha curcas] |
| 18 |
Hb_017159_010 |
0.2055083547 |
- |
- |
- |
| 19 |
Hb_002311_110 |
0.2105158314 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000649_270 |
0.2106214457 |
- |
- |
DUF26 domain-containing protein 1 precursor, putative [Ricinus communis] |