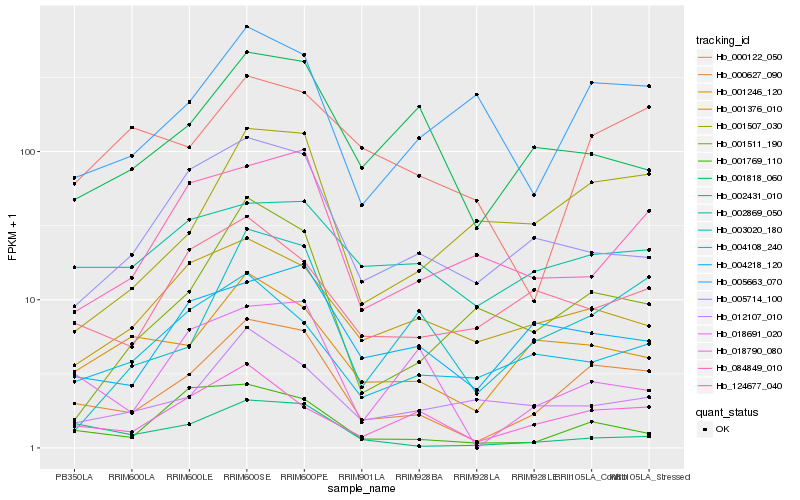
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000627_090 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001246_120 |
0.1926558996 |
- |
- |
PREDICTED: uncharacterized protein LOC105643270 [Jatropha curcas] |
| 3 |
Hb_018790_080 |
0.1927016541 |
- |
- |
hypothetical protein POPTR_0011s06335g [Populus trichocarpa] |
| 4 |
Hb_004218_120 |
0.2006285391 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase SINAT5-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_001507_030 |
0.2034026748 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 2 [Jatropha curcas] |
| 6 |
Hb_003020_180 |
0.2039135568 |
- |
- |
PREDICTED: probable protein phosphatase 2C 24 [Jatropha curcas] |
| 7 |
Hb_002431_010 |
0.2090319352 |
- |
- |
PREDICTED: uncharacterized protein LOC104599557 isoform X2 [Nelumbo nucifera] |
| 8 |
Hb_005663_070 |
0.2103687267 |
- |
- |
dehydrin protein [Manihot esculenta] |
| 9 |
Hb_001769_110 |
0.2120653425 |
- |
- |
PREDICTED: probable metal-nicotianamine transporter YSL7 [Jatropha curcas] |
| 10 |
Hb_005714_100 |
0.2122195412 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Vitis vinifera] |
| 11 |
Hb_002869_050 |
0.213974263 |
- |
- |
PREDICTED: transmembrane and coiled-coil domain-containing protein 1 [Jatropha curcas] |
| 12 |
Hb_124677_040 |
0.2155075774 |
- |
- |
PREDICTED: uncharacterized protein LOC105650471 [Jatropha curcas] |
| 13 |
Hb_004108_240 |
0.2159112428 |
- |
- |
hypothetical protein POPTR_0002s05550g [Populus trichocarpa] |
| 14 |
Hb_001376_010 |
0.2168986306 |
- |
- |
diacylglycerol kinase, alpha, putative [Ricinus communis] |
| 15 |
Hb_001818_060 |
0.2189071816 |
- |
- |
PREDICTED: cysteine proteinase inhibitor 5-like [Jatropha curcas] |
| 16 |
Hb_012107_010 |
0.2202008827 |
- |
- |
hypothetical protein JCGZ_19531 [Jatropha curcas] |
| 17 |
Hb_001511_190 |
0.2203069912 |
- |
- |
PREDICTED: probable protein phosphatase 2C 25 [Jatropha curcas] |
| 18 |
Hb_000122_050 |
0.220404525 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_084849_010 |
0.221432265 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_018691_020 |
0.2215065198 |
- |
- |
- |