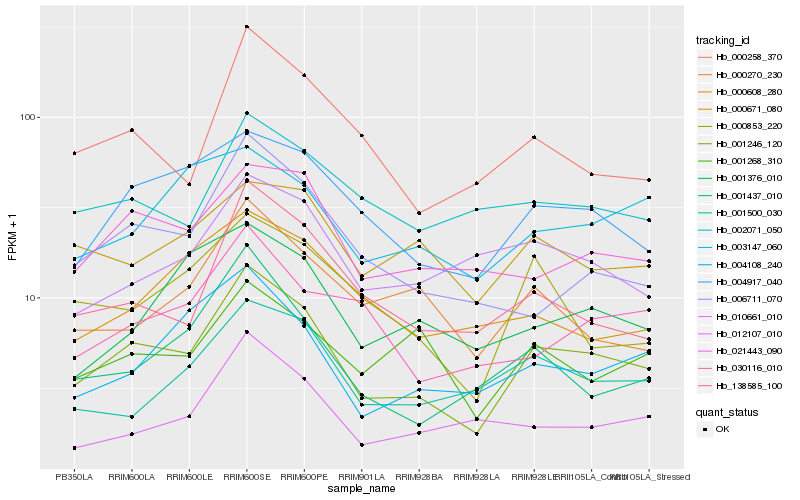
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001246_120 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105643270 [Jatropha curcas] |
| 2 |
Hb_004917_040 |
0.1342322082 |
- |
- |
hypothetical protein POPTR_0009s03470g [Populus trichocarpa] |
| 3 |
Hb_021443_090 |
0.1454812463 |
- |
- |
PREDICTED: xyloglucan galactosyltransferase KATAMARI1 homolog [Jatropha curcas] |
| 4 |
Hb_000258_370 |
0.1462231859 |
- |
- |
hypothetical protein [Ricinus communis] |
| 5 |
Hb_138585_100 |
0.1484546213 |
- |
- |
PREDICTED: uncharacterized protein LOC105648740 [Jatropha curcas] |
| 6 |
Hb_006711_070 |
0.1495690648 |
- |
- |
hypothetical protein RCOM_1336320 [Ricinus communis] |
| 7 |
Hb_004108_240 |
0.1515883111 |
- |
- |
hypothetical protein POPTR_0002s05550g [Populus trichocarpa] |
| 8 |
Hb_001437_010 |
0.1527690593 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g33260 [Jatropha curcas] |
| 9 |
Hb_001500_030 |
0.1571406945 |
- |
- |
PREDICTED: uncharacterized protein At2g33490 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000853_220 |
0.1607715141 |
- |
- |
PREDICTED: cytidine deaminase 1 [Jatropha curcas] |
| 11 |
Hb_002071_050 |
0.1613755875 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 5-like [Jatropha curcas] |
| 12 |
Hb_001268_310 |
0.1629970109 |
- |
- |
- |
| 13 |
Hb_012107_010 |
0.1647003141 |
- |
- |
hypothetical protein JCGZ_19531 [Jatropha curcas] |
| 14 |
Hb_030116_010 |
0.1647519846 |
- |
- |
F-box protein, atfbl3, putative [Ricinus communis] |
| 15 |
Hb_000270_230 |
0.1656636083 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 19 [Jatropha curcas] |
| 16 |
Hb_010661_010 |
0.1663187703 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX9H isoform X1 [Jatropha curcas] |
| 17 |
Hb_000671_080 |
0.1670804331 |
- |
- |
PREDICTED: 5'-adenylylsulfate reductase-like 4 [Jatropha curcas] |
| 18 |
Hb_001376_010 |
0.1687591717 |
- |
- |
diacylglycerol kinase, alpha, putative [Ricinus communis] |
| 19 |
Hb_003147_060 |
0.1696496279 |
- |
- |
syntaxin, plant, putative [Ricinus communis] |
| 20 |
Hb_000608_280 |
0.1727011774 |
- |
- |
Aspartic proteinase Asp1 precursor, putative [Ricinus communis] |