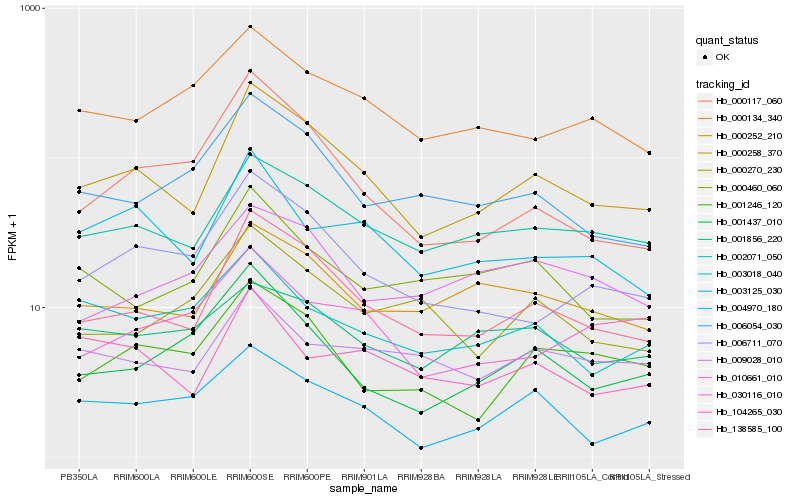
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000258_370 |
0.0 |
- |
- |
hypothetical protein [Ricinus communis] |
| 2 |
Hb_138585_100 |
0.0551278054 |
- |
- |
PREDICTED: uncharacterized protein LOC105648740 [Jatropha curcas] |
| 3 |
Hb_002071_050 |
0.1121311785 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 5-like [Jatropha curcas] |
| 4 |
Hb_006711_070 |
0.1278075735 |
- |
- |
hypothetical protein RCOM_1336320 [Ricinus communis] |
| 5 |
Hb_000252_210 |
0.1337860891 |
- |
- |
PREDICTED: uncharacterized protein LOC105644655 [Jatropha curcas] |
| 6 |
Hb_003125_030 |
0.1380165636 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 7 |
Hb_006054_030 |
0.1410591638 |
- |
- |
PREDICTED: protein GPR107-like [Jatropha curcas] |
| 8 |
Hb_000117_060 |
0.1441350022 |
- |
- |
PREDICTED: uncharacterized protein LOC103949199 [Pyrus x bretschneideri] |
| 9 |
Hb_001246_120 |
0.1462231859 |
- |
- |
PREDICTED: uncharacterized protein LOC105643270 [Jatropha curcas] |
| 10 |
Hb_000134_340 |
0.1489897662 |
- |
- |
PREDICTED: zinc finger A20 and AN1 domain-containing stress-associated protein 5 [Jatropha curcas] |
| 11 |
Hb_104265_030 |
0.1550115726 |
- |
- |
hypothetical protein POPTR_0001s31230g [Populus trichocarpa] |
| 12 |
Hb_000460_060 |
0.1564634796 |
- |
- |
importin beta-1, putative [Ricinus communis] |
| 13 |
Hb_009028_010 |
0.1573008268 |
- |
- |
PREDICTED: probable disease resistance protein At1g12280 [Jatropha curcas] |
| 14 |
Hb_004970_180 |
0.1577331224 |
- |
- |
- |
| 15 |
Hb_001437_010 |
0.1581831988 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g33260 [Jatropha curcas] |
| 16 |
Hb_000270_230 |
0.160532769 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 19 [Jatropha curcas] |
| 17 |
Hb_003018_040 |
0.1629848485 |
transcription factor |
TF Family: C3H |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001856_220 |
0.163847242 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_010661_010 |
0.1649227293 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX9H isoform X1 [Jatropha curcas] |
| 20 |
Hb_030116_010 |
0.1649419157 |
- |
- |
F-box protein, atfbl3, putative [Ricinus communis] |