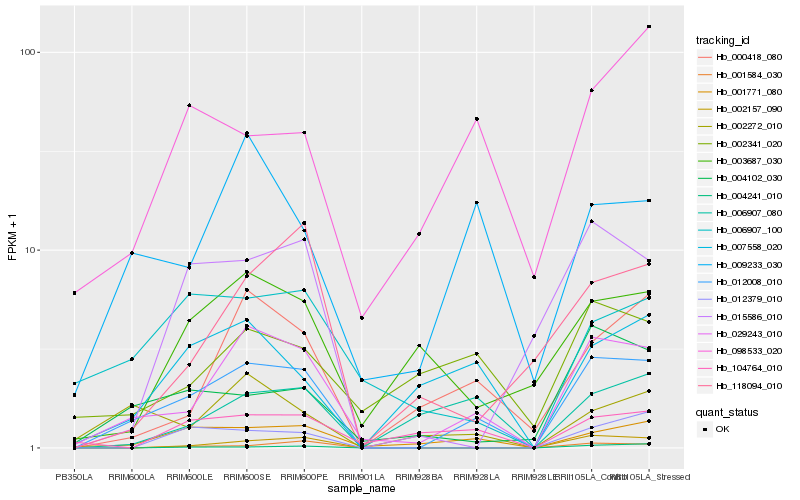
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012008_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_029243_010 |
0.1433934261 |
- |
- |
PREDICTED: protein LURP-one-related 15-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_001771_080 |
0.2214051657 |
- |
- |
PREDICTED: UDP-D-apiose/UDP-D-xylose synthase 2 [Amborella trichopoda] |
| 4 |
Hb_002157_090 |
0.2521682815 |
- |
- |
hypothetical protein VITISV_041741 [Vitis vinifera] |
| 5 |
Hb_004102_030 |
0.2572385061 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At5g63020 [Jatropha curcas] |
| 6 |
Hb_007558_020 |
0.2621107067 |
- |
- |
hypothetical protein JCGZ_17631 [Jatropha curcas] |
| 7 |
Hb_000418_080 |
0.2621755023 |
- |
- |
PREDICTED: uncharacterized protein LOC105124652 [Populus euphratica] |
| 8 |
Hb_009233_030 |
0.2689580151 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 9 |
Hb_002272_010 |
0.2740430162 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_012379_010 |
0.2764799077 |
- |
- |
- |
| 11 |
Hb_015586_010 |
0.2774419815 |
- |
- |
PREDICTED: flavonol sulfotransferase-like [Jatropha curcas] |
| 12 |
Hb_006907_080 |
0.2832762843 |
- |
- |
PREDICTED: RPM1-interacting protein 4-like [Jatropha curcas] |
| 13 |
Hb_001584_030 |
0.2869684369 |
- |
- |
PREDICTED: glycerophosphodiester phosphodiesterase protein kinase domain-containing GDPDL2-like [Nelumbo nucifera] |
| 14 |
Hb_098533_020 |
0.2892737479 |
- |
- |
PREDICTED: zinc finger AN1 domain-containing stress-associated protein 12 [Jatropha curcas] |
| 15 |
Hb_006907_100 |
0.2894673365 |
- |
- |
PREDICTED: tropinone reductase-like 3 [Jatropha curcas] |
| 16 |
Hb_004241_010 |
0.2903248579 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 17 |
Hb_003687_030 |
0.2959364182 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 18 |
Hb_104764_010 |
0.2980854657 |
- |
- |
hypothetical protein JCGZ_16024 [Jatropha curcas] |
| 19 |
Hb_118094_010 |
0.2982916995 |
- |
- |
hypothetical protein POPTR_0011s06595g [Populus trichocarpa] |
| 20 |
Hb_002341_020 |
0.2992059892 |
- |
- |
PREDICTED: pyruvate decarboxylase 2 [Populus euphratica] |