| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
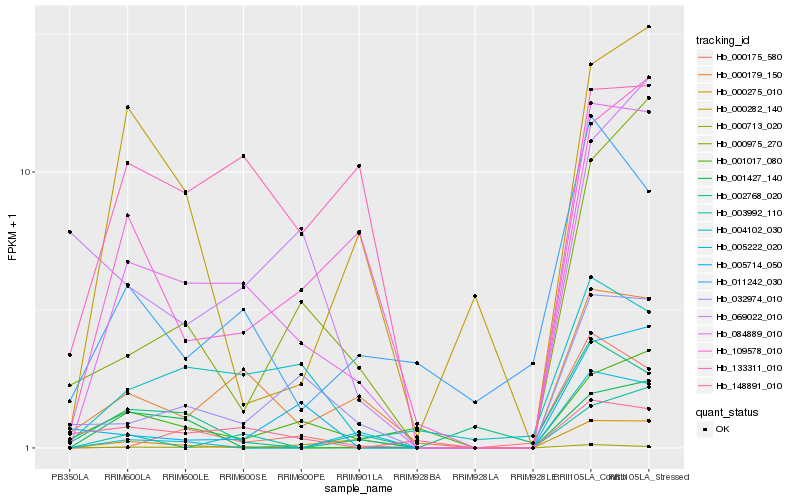
Hb_084889_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638918 [Jatropha curcas] |
| 2 |
Hb_000179_150 |
0.1828923514 |
- |
- |
hypothetical protein ZEAMMB73_153699 [Zea mays] |
| 3 |
Hb_109578_010 |
0.2219457649 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_032974_010 |
0.2235776845 |
- |
- |
- |
| 5 |
Hb_004102_030 |
0.2317810234 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At5g63020 [Jatropha curcas] |
| 6 |
Hb_000975_270 |
0.2365779692 |
- |
- |
PREDICTED: histone H2B.11-like [Phoenix dactylifera] |
| 7 |
Hb_001017_080 |
0.2544254062 |
- |
- |
hypothetical protein POPTR_0010s06020g [Populus trichocarpa] |
| 8 |
Hb_000275_010 |
0.2555902573 |
- |
- |
- |
| 9 |
Hb_011242_030 |
0.2601670562 |
- |
- |
- |
| 10 |
Hb_000175_580 |
0.2639012887 |
transcription factor, desease resistance |
TF Family: C3H, Gene Name: zf-CCCH |
hypothetical protein POPTR_0013s00890g [Populus trichocarpa] |
| 11 |
Hb_000713_020 |
0.2671708319 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 12 |
Hb_133311_010 |
0.2691763481 |
- |
- |
PREDICTED: U-box domain-containing protein 62-like isoform X1 [Glycine max] |
| 13 |
Hb_000282_140 |
0.2719878116 |
- |
- |
hypothetical protein JCGZ_07670 [Jatropha curcas] |
| 14 |
Hb_003992_110 |
0.2760628049 |
- |
- |
hypothetical protein VITISV_016971 [Vitis vinifera] |
| 15 |
Hb_002768_020 |
0.2843159927 |
transcription factor |
TF Family: MYB |
hypothetical protein JCGZ_00902 [Jatropha curcas] |
| 16 |
Hb_005222_020 |
0.2843336506 |
- |
- |
hypothetical protein [Beta vulgaris subsp. vulgaris] |
| 17 |
Hb_005714_050 |
0.2895049731 |
transcription factor |
TF Family: B3 |
hypothetical protein POPTR_1461s00200g [Populus trichocarpa] |
| 18 |
Hb_001427_140 |
0.2897475274 |
- |
- |
PREDICTED: probable phospholipase A2 homolog 1 [Jatropha curcas] |
| 19 |
Hb_069022_010 |
0.2916267539 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_148891_010 |
0.2922328302 |
- |
- |
- |