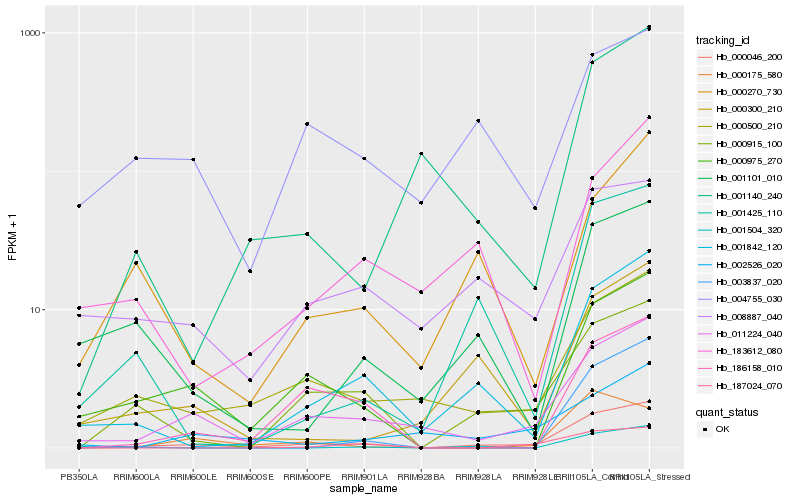
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000046_200 |
0.0 |
- |
- |
PREDICTED: bifunctional 3-dehydroquinate dehydratase/shikimate dehydrogenase, chloroplastic-like [Jatropha curcas] |
| 2 |
Hb_011224_040 |
0.164584111 |
- |
- |
PREDICTED: ER membrane protein complex subunit 8/9 homolog [Jatropha curcas] |
| 3 |
Hb_000915_100 |
0.1736439016 |
- |
- |
PREDICTED: auxin-induced protein 15A [Jatropha curcas] |
| 4 |
Hb_001842_120 |
0.1829806177 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 5 |
Hb_000500_210 |
0.1987087006 |
- |
- |
- |
| 6 |
Hb_000975_270 |
0.2169890483 |
- |
- |
PREDICTED: histone H2B.11-like [Phoenix dactylifera] |
| 7 |
Hb_001425_110 |
0.2191676804 |
- |
- |
hypothetical protein Csa_6G106200 [Cucumis sativus] |
| 8 |
Hb_186158_010 |
0.2206606716 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000300_210 |
0.2285439196 |
- |
- |
gd2b, putative [Ricinus communis] |
| 10 |
Hb_187024_070 |
0.234149126 |
- |
- |
PREDICTED: uncharacterized protein LOC105785446 [Gossypium raimondii] |
| 11 |
Hb_001140_240 |
0.2350142725 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 12 |
Hb_001101_010 |
0.2390097549 |
- |
- |
hypothetical protein POPTR_0004s18730g [Populus trichocarpa] |
| 13 |
Hb_008887_040 |
0.2419179876 |
- |
- |
Ribonuclease P family protein / Rpp14 family protein isoform 1 [Theobroma cacao] |
| 14 |
Hb_000270_730 |
0.243158266 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2-23 kDa-like [Jatropha curcas] |
| 15 |
Hb_002526_020 |
0.2445486289 |
- |
- |
- |
| 16 |
Hb_001504_320 |
0.2448797711 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105634775 [Jatropha curcas] |
| 17 |
Hb_003837_020 |
0.2471789673 |
- |
- |
PREDICTED: uncharacterized protein LOC103710465 isoform X1 [Phoenix dactylifera] |
| 18 |
Hb_000175_580 |
0.25106096 |
transcription factor, desease resistance |
TF Family: C3H, Gene Name: zf-CCCH |
hypothetical protein POPTR_0013s00890g [Populus trichocarpa] |
| 19 |
Hb_183612_080 |
0.2547789095 |
- |
- |
hypothetical protein POPTR_0001s00500g [Populus trichocarpa] |
| 20 |
Hb_004755_030 |
0.2550435839 |
- |
- |
EG964 [Manihot glaziovii] |