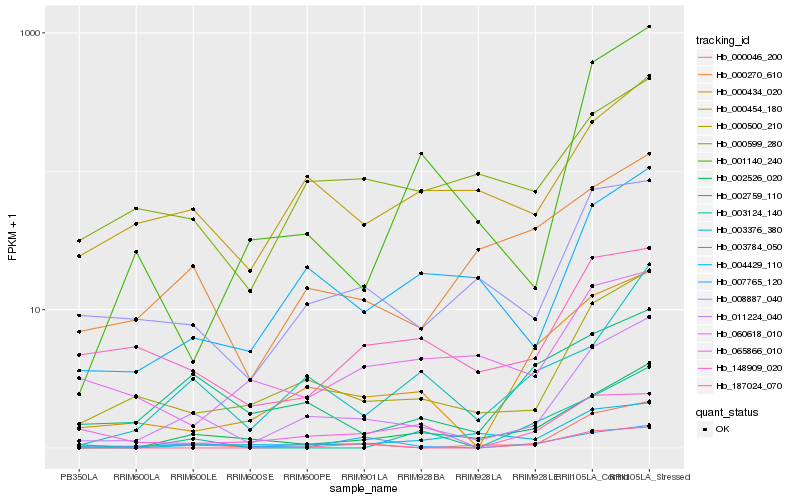
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000434_020 |
0.0 |
- |
- |
PREDICTED: protein yippee-like At5g53940 [Jatropha curcas] |
| 2 |
Hb_011224_040 |
0.1993156964 |
- |
- |
PREDICTED: ER membrane protein complex subunit 8/9 homolog [Jatropha curcas] |
| 3 |
Hb_000500_210 |
0.2046494034 |
- |
- |
- |
| 4 |
Hb_002526_020 |
0.2215955137 |
- |
- |
- |
| 5 |
Hb_065866_010 |
0.2224568426 |
- |
- |
PREDICTED: hydroxyacyl-thioester dehydratase type 2, mitochondrial [Fragaria vesca subsp. vesca] |
| 6 |
Hb_002759_110 |
0.2315103886 |
- |
- |
RNA-binding family protein, putative isoform 1 [Theobroma cacao] |
| 7 |
Hb_187024_070 |
0.2467255921 |
- |
- |
PREDICTED: uncharacterized protein LOC105785446 [Gossypium raimondii] |
| 8 |
Hb_000270_610 |
0.2525488565 |
- |
- |
PREDICTED: uncharacterized protein At5g50100, mitochondrial [Jatropha curcas] |
| 9 |
Hb_000046_200 |
0.2560687823 |
- |
- |
PREDICTED: bifunctional 3-dehydroquinate dehydratase/shikimate dehydrogenase, chloroplastic-like [Jatropha curcas] |
| 10 |
Hb_003376_380 |
0.2564679828 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG3 [Jatropha curcas] |
| 11 |
Hb_148909_020 |
0.2590214717 |
- |
- |
PREDICTED: glutamyl-tRNA(Gln) amidotransferase subunit C, chloroplastic/mitochondrial [Jatropha curcas] |
| 12 |
Hb_003124_140 |
0.2625765989 |
- |
- |
hypothetical protein Csa_4G182240 [Cucumis sativus] |
| 13 |
Hb_000454_180 |
0.2646748872 |
- |
- |
PREDICTED: uncharacterized protein LOC105134496 [Populus euphratica] |
| 14 |
Hb_003784_050 |
0.2647473486 |
- |
- |
PREDICTED: replication protein A 32 kDa subunit B [Jatropha curcas] |
| 15 |
Hb_008887_040 |
0.264766529 |
- |
- |
Ribonuclease P family protein / Rpp14 family protein isoform 1 [Theobroma cacao] |
| 16 |
Hb_060618_010 |
0.2662611683 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 17 |
Hb_004429_110 |
0.2688750108 |
- |
- |
PREDICTED: protein SPIRAL1-like 1 [Populus euphratica] |
| 18 |
Hb_007765_120 |
0.2692874661 |
- |
- |
glycogen phosphorylase, putative [Ricinus communis] |
| 19 |
Hb_000599_280 |
0.271272754 |
- |
- |
PREDICTED: 60S ribosomal protein L14-1 [Jatropha curcas] |
| 20 |
Hb_001140_240 |
0.2717278976 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |