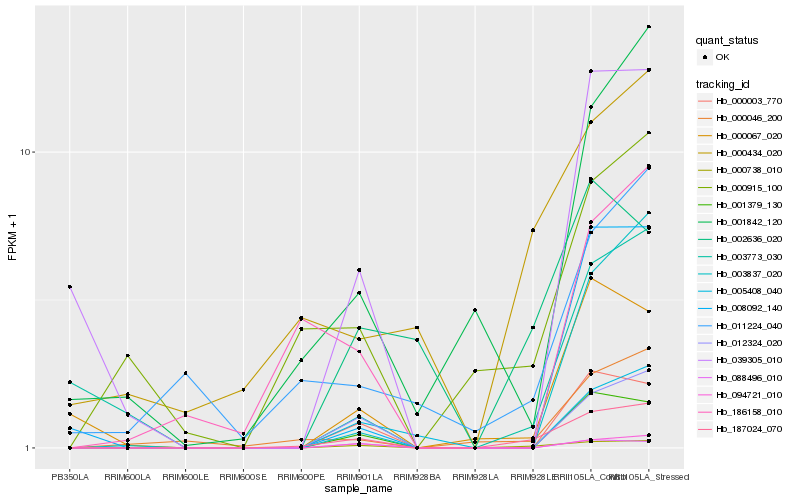
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_187024_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105785446 [Gossypium raimondii] |
| 2 |
Hb_000738_010 |
0.1229802948 |
- |
- |
hypothetical protein PRUPE_ppa021229mg [Prunus persica] |
| 3 |
Hb_000046_200 |
0.234149126 |
- |
- |
PREDICTED: bifunctional 3-dehydroquinate dehydratase/shikimate dehydrogenase, chloroplastic-like [Jatropha curcas] |
| 4 |
Hb_001379_130 |
0.2350900274 |
- |
- |
PREDICTED: pre-mRNA-processing protein 40A isoform X2 [Jatropha curcas] |
| 5 |
Hb_012324_020 |
0.2368047253 |
- |
- |
- |
| 6 |
Hb_094721_010 |
0.2380391118 |
- |
- |
PREDICTED: uncharacterized protein LOC104809818, partial [Tarenaya hassleriana] |
| 7 |
Hb_000003_770 |
0.239080852 |
- |
- |
- |
| 8 |
Hb_000434_020 |
0.2467255921 |
- |
- |
PREDICTED: protein yippee-like At5g53940 [Jatropha curcas] |
| 9 |
Hb_000915_100 |
0.2492975337 |
- |
- |
PREDICTED: auxin-induced protein 15A [Jatropha curcas] |
| 10 |
Hb_002636_020 |
0.2680756321 |
- |
- |
- |
| 11 |
Hb_001842_120 |
0.270092874 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 12 |
Hb_003837_020 |
0.2715807367 |
- |
- |
PREDICTED: uncharacterized protein LOC103710465 isoform X1 [Phoenix dactylifera] |
| 13 |
Hb_000067_020 |
0.2731360404 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: inactive disease resistance protein RPS4-like [Jatropha curcas] |
| 14 |
Hb_008092_140 |
0.2759585491 |
- |
- |
PREDICTED: uncharacterized protein LOC103636948 [Zea mays] |
| 15 |
Hb_011224_040 |
0.2764175991 |
- |
- |
PREDICTED: ER membrane protein complex subunit 8/9 homolog [Jatropha curcas] |
| 16 |
Hb_005408_040 |
0.2777036479 |
- |
- |
- |
| 17 |
Hb_088496_010 |
0.2797280867 |
- |
- |
PREDICTED: uncharacterized protein LOC104889111 [Beta vulgaris subsp. vulgaris] |
| 18 |
Hb_039305_010 |
0.2842253427 |
- |
- |
hypothetical protein JCGZ_18422 [Jatropha curcas] |
| 19 |
Hb_003773_030 |
0.2870191606 |
- |
- |
- |
| 20 |
Hb_186158_010 |
0.2881898507 |
- |
- |
conserved hypothetical protein [Ricinus communis] |