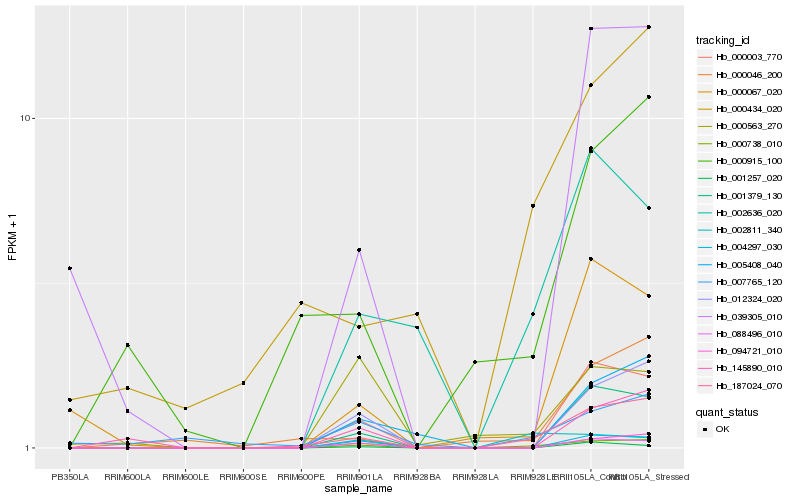
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000738_010 |
0.0 |
- |
- |
hypothetical protein PRUPE_ppa021229mg [Prunus persica] |
| 2 |
Hb_187024_070 |
0.1229802948 |
- |
- |
PREDICTED: uncharacterized protein LOC105785446 [Gossypium raimondii] |
| 3 |
Hb_012324_020 |
0.232651998 |
- |
- |
- |
| 4 |
Hb_094721_010 |
0.2328795912 |
- |
- |
PREDICTED: uncharacterized protein LOC104809818, partial [Tarenaya hassleriana] |
| 5 |
Hb_000003_770 |
0.2413353545 |
- |
- |
- |
| 6 |
Hb_002636_020 |
0.2426217378 |
- |
- |
- |
| 7 |
Hb_001379_130 |
0.2481136477 |
- |
- |
PREDICTED: pre-mRNA-processing protein 40A isoform X2 [Jatropha curcas] |
| 8 |
Hb_088496_010 |
0.2500118965 |
- |
- |
PREDICTED: uncharacterized protein LOC104889111 [Beta vulgaris subsp. vulgaris] |
| 9 |
Hb_002811_340 |
0.2563185126 |
- |
- |
PREDICTED: ammonium transporter 3 member 2-like [Populus euphratica] |
| 10 |
Hb_004297_030 |
0.2600217235 |
- |
- |
PREDICTED: uncharacterized protein LOC105633198 [Jatropha curcas] |
| 11 |
Hb_005408_040 |
0.2852377331 |
- |
- |
- |
| 12 |
Hb_000067_020 |
0.2889573312 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: inactive disease resistance protein RPS4-like [Jatropha curcas] |
| 13 |
Hb_145890_010 |
0.291374499 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 14 |
Hb_000563_270 |
0.2933943364 |
- |
- |
- |
| 15 |
Hb_000434_020 |
0.3027178788 |
- |
- |
PREDICTED: protein yippee-like At5g53940 [Jatropha curcas] |
| 16 |
Hb_007765_120 |
0.3061822881 |
- |
- |
glycogen phosphorylase, putative [Ricinus communis] |
| 17 |
Hb_039305_010 |
0.3063363452 |
- |
- |
hypothetical protein JCGZ_18422 [Jatropha curcas] |
| 18 |
Hb_001257_020 |
0.3067852315 |
- |
- |
- |
| 19 |
Hb_000915_100 |
0.3114574839 |
- |
- |
PREDICTED: auxin-induced protein 15A [Jatropha curcas] |
| 20 |
Hb_000046_200 |
0.3118002619 |
- |
- |
PREDICTED: bifunctional 3-dehydroquinate dehydratase/shikimate dehydrogenase, chloroplastic-like [Jatropha curcas] |