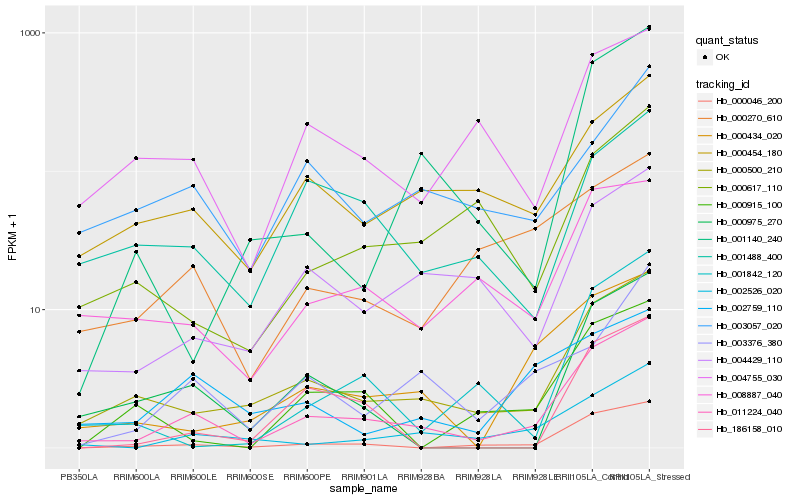
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011224_040 |
0.0 |
- |
- |
PREDICTED: ER membrane protein complex subunit 8/9 homolog [Jatropha curcas] |
| 2 |
Hb_000500_210 |
0.1484417309 |
- |
- |
- |
| 3 |
Hb_000046_200 |
0.164584111 |
- |
- |
PREDICTED: bifunctional 3-dehydroquinate dehydratase/shikimate dehydrogenase, chloroplastic-like [Jatropha curcas] |
| 4 |
Hb_002526_020 |
0.1807691303 |
- |
- |
- |
| 5 |
Hb_000434_020 |
0.1993156964 |
- |
- |
PREDICTED: protein yippee-like At5g53940 [Jatropha curcas] |
| 6 |
Hb_004429_110 |
0.2023807344 |
- |
- |
PREDICTED: protein SPIRAL1-like 1 [Populus euphratica] |
| 7 |
Hb_000975_270 |
0.2078183118 |
- |
- |
PREDICTED: histone H2B.11-like [Phoenix dactylifera] |
| 8 |
Hb_008887_040 |
0.2087914703 |
- |
- |
Ribonuclease P family protein / Rpp14 family protein isoform 1 [Theobroma cacao] |
| 9 |
Hb_000454_180 |
0.2101133578 |
- |
- |
PREDICTED: uncharacterized protein LOC105134496 [Populus euphratica] |
| 10 |
Hb_003376_380 |
0.2124635494 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG3 [Jatropha curcas] |
| 11 |
Hb_004755_030 |
0.2191889434 |
- |
- |
EG964 [Manihot glaziovii] |
| 12 |
Hb_001842_120 |
0.2242390695 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 13 |
Hb_000617_110 |
0.2290532949 |
- |
- |
translationally controlled tumor protein [Hevea brasiliensis] |
| 14 |
Hb_000270_610 |
0.2304670045 |
- |
- |
PREDICTED: uncharacterized protein At5g50100, mitochondrial [Jatropha curcas] |
| 15 |
Hb_001140_240 |
0.2345997099 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 16 |
Hb_002759_110 |
0.2361954318 |
- |
- |
RNA-binding family protein, putative isoform 1 [Theobroma cacao] |
| 17 |
Hb_186158_010 |
0.2362017602 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001488_400 |
0.2368257504 |
- |
- |
hypothetical protein POPTR_0018s12030g [Populus trichocarpa] |
| 19 |
Hb_000915_100 |
0.2369709489 |
- |
- |
PREDICTED: auxin-induced protein 15A [Jatropha curcas] |
| 20 |
Hb_003057_020 |
0.239005182 |
- |
- |
40S ribosomal protein S23, putative [Ricinus communis] |