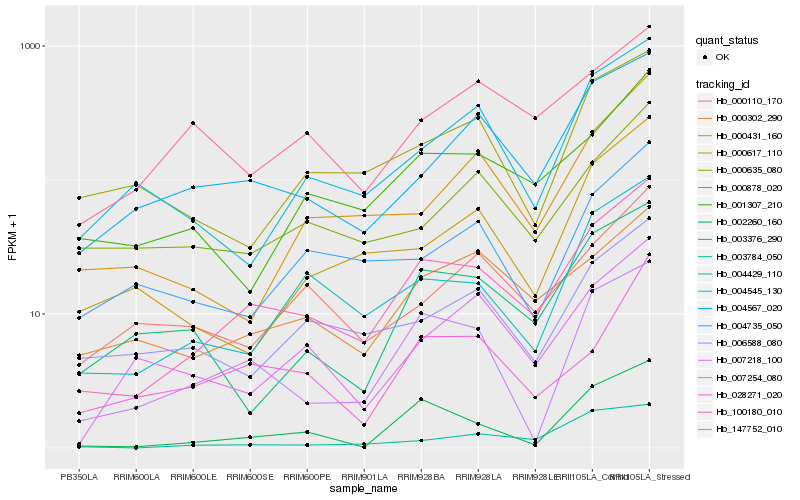
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_100180_010 |
0.0 |
- |
- |
PREDICTED: disease resistance protein RPM1-like [Jatropha curcas] |
| 2 |
Hb_004429_110 |
0.1438601078 |
- |
- |
PREDICTED: protein SPIRAL1-like 1 [Populus euphratica] |
| 3 |
Hb_004567_020 |
0.1506022605 |
- |
- |
Small ubiquitin-related modifier 2 [Morus notabilis] |
| 4 |
Hb_001307_210 |
0.1569158812 |
- |
- |
Os02g0814700 [Oryza sativa Japonica Group] |
| 5 |
Hb_007218_100 |
0.1592573992 |
- |
- |
phosphofructokinase [Hevea brasiliensis] |
| 6 |
Hb_000302_290 |
0.1632656226 |
- |
- |
40S ribosomal S10-3 -like protein [Gossypium arboreum] |
| 7 |
Hb_002260_160 |
0.1659771817 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_004545_130 |
0.1696343425 |
- |
- |
uncharacterized protein LOC100305465 [Glycine max] |
| 9 |
Hb_003784_050 |
0.1753945759 |
- |
- |
PREDICTED: replication protein A 32 kDa subunit B [Jatropha curcas] |
| 10 |
Hb_006588_080 |
0.1755278642 |
- |
- |
hypothetical protein JCGZ_25746 [Jatropha curcas] |
| 11 |
Hb_000110_170 |
0.1756078322 |
- |
- |
PREDICTED: arginine biosynthesis bifunctional protein ArgJ, chloroplastic isoform X1 [Jatropha curcas] |
| 12 |
Hb_028271_020 |
0.1771396398 |
- |
- |
acyl-CoA oxidase, putative [Ricinus communis] |
| 13 |
Hb_003376_290 |
0.1780483322 |
- |
- |
hypothetical protein POPTR_0010s22880g [Populus trichocarpa] |
| 14 |
Hb_004735_050 |
0.1782269059 |
- |
- |
PREDICTED: uncharacterized protein LOC105639312 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000617_110 |
0.178511986 |
- |
- |
translationally controlled tumor protein [Hevea brasiliensis] |
| 16 |
Hb_000431_160 |
0.1786565324 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit gamma-like [Jatropha curcas] |
| 17 |
Hb_007254_080 |
0.1789135916 |
- |
- |
- |
| 18 |
Hb_000878_020 |
0.1795877299 |
- |
- |
PREDICTED: uncharacterized protein LOC105629937 [Jatropha curcas] |
| 19 |
Hb_147752_010 |
0.18106529 |
- |
- |
PREDICTED: 40S ribosomal protein S9-2-like isoform X1 [Populus euphratica] |
| 20 |
Hb_000635_080 |
0.1818816261 |
- |
- |
60S ribosomal protein L17A [Hevea brasiliensis] |