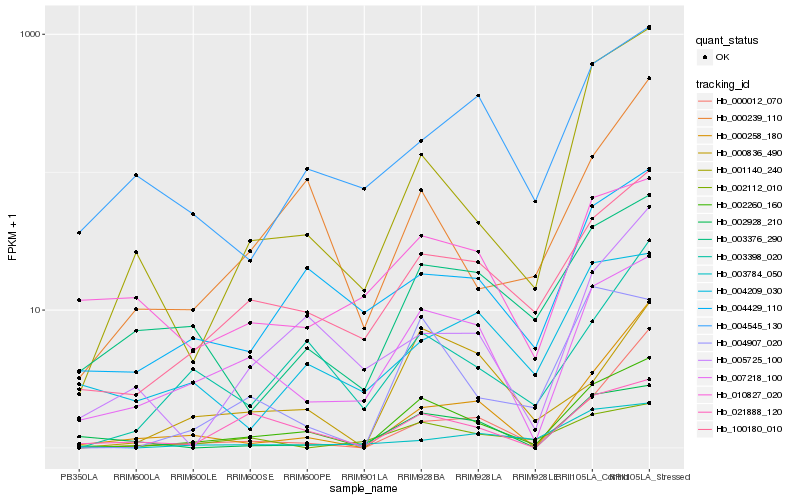
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002260_160 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_100180_010 |
0.1659771817 |
- |
- |
PREDICTED: disease resistance protein RPM1-like [Jatropha curcas] |
| 3 |
Hb_007218_100 |
0.1697395535 |
- |
- |
phosphofructokinase [Hevea brasiliensis] |
| 4 |
Hb_004429_110 |
0.1974318428 |
- |
- |
PREDICTED: protein SPIRAL1-like 1 [Populus euphratica] |
| 5 |
Hb_004907_020 |
0.204655722 |
- |
- |
PREDICTED: ABC transporter G family member 6-like [Jatropha curcas] |
| 6 |
Hb_021888_120 |
0.2085533247 |
- |
- |
PREDICTED: putative CCR4-associated factor 1 homolog 8 [Jatropha curcas] |
| 7 |
Hb_001140_240 |
0.2087682009 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 8 |
Hb_003398_020 |
0.2145883324 |
- |
- |
PREDICTED: uncharacterized protein LOC105647439 [Jatropha curcas] |
| 9 |
Hb_003376_290 |
0.2157459063 |
- |
- |
hypothetical protein POPTR_0010s22880g [Populus trichocarpa] |
| 10 |
Hb_000239_110 |
0.2232102115 |
- |
- |
PREDICTED: adenine phosphoribosyltransferase 1-like [Jatropha curcas] |
| 11 |
Hb_002928_210 |
0.2275172339 |
- |
- |
- |
| 12 |
Hb_005725_100 |
0.2306995761 |
- |
- |
hypothetical protein 30 [Hevea brasiliensis] |
| 13 |
Hb_010827_020 |
0.2385410812 |
- |
- |
60S ribosomal protein L17B [Hevea brasiliensis] |
| 14 |
Hb_004545_130 |
0.2391867925 |
- |
- |
uncharacterized protein LOC100305465 [Glycine max] |
| 15 |
Hb_000258_180 |
0.239301463 |
- |
- |
PREDICTED: glutathione S-transferase U10-like [Vitis vinifera] |
| 16 |
Hb_000012_070 |
0.2400070495 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 17 |
Hb_000836_490 |
0.2405396315 |
- |
- |
PREDICTED: E3 SUMO-protein ligase pli1-like [Jatropha curcas] |
| 18 |
Hb_004209_030 |
0.2433189345 |
- |
- |
PREDICTED: (DL)-glycerol-3-phosphatase 2 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002112_010 |
0.2459416891 |
- |
- |
hypothetical protein POPTR_0001s45830g [Populus trichocarpa] |
| 20 |
Hb_003784_050 |
0.2472937228 |
- |
- |
PREDICTED: replication protein A 32 kDa subunit B [Jatropha curcas] |