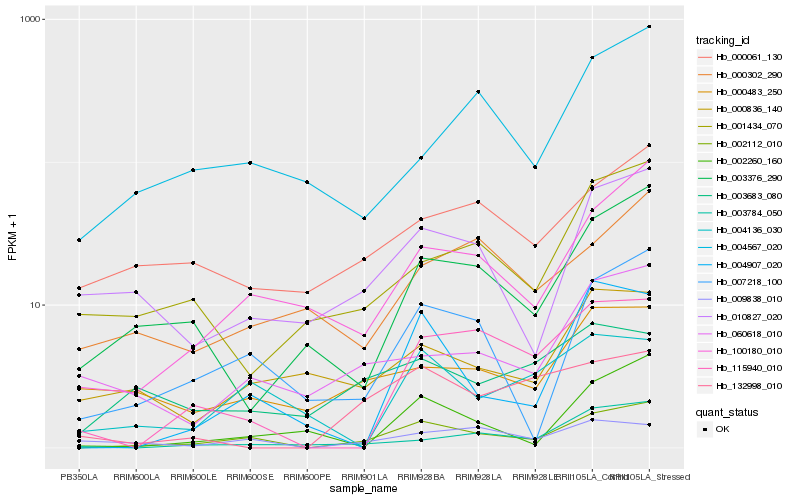
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002112_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0001s45830g [Populus trichocarpa] |
| 2 |
Hb_100180_010 |
0.189472305 |
- |
- |
PREDICTED: disease resistance protein RPM1-like [Jatropha curcas] |
| 3 |
Hb_007218_100 |
0.1939260145 |
- |
- |
phosphofructokinase [Hevea brasiliensis] |
| 4 |
Hb_010827_020 |
0.2293866175 |
- |
- |
60S ribosomal protein L17B [Hevea brasiliensis] |
| 5 |
Hb_004907_020 |
0.2359304464 |
- |
- |
PREDICTED: ABC transporter G family member 6-like [Jatropha curcas] |
| 6 |
Hb_003376_290 |
0.2371623054 |
- |
- |
hypothetical protein POPTR_0010s22880g [Populus trichocarpa] |
| 7 |
Hb_060618_010 |
0.2404876215 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 8 |
Hb_115940_010 |
0.2430048021 |
- |
- |
putative auxin-amidohydrolase precursor family protein [Populus trichocarpa] |
| 9 |
Hb_003784_050 |
0.2450175387 |
- |
- |
PREDICTED: replication protein A 32 kDa subunit B [Jatropha curcas] |
| 10 |
Hb_000836_140 |
0.2456231809 |
- |
- |
PREDICTED: protein TIC 20-IV, chloroplastic [Jatropha curcas] |
| 11 |
Hb_002260_160 |
0.2459416891 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000061_130 |
0.2464050407 |
- |
- |
rotamase, putative [Ricinus communis] |
| 13 |
Hb_004567_020 |
0.2470919172 |
- |
- |
Small ubiquitin-related modifier 2 [Morus notabilis] |
| 14 |
Hb_009838_010 |
0.2489579541 |
- |
- |
- |
| 15 |
Hb_001434_070 |
0.2497371423 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM8 [Jatropha curcas] |
| 16 |
Hb_000483_250 |
0.2524748458 |
- |
- |
PREDICTED: uncharacterized protein LOC105803540 [Gossypium raimondii] |
| 17 |
Hb_000302_290 |
0.2571055585 |
- |
- |
40S ribosomal S10-3 -like protein [Gossypium arboreum] |
| 18 |
Hb_004136_030 |
0.2578307341 |
- |
- |
- |
| 19 |
Hb_003683_080 |
0.2606151177 |
- |
- |
Angio-associated migratory cell protein, putative [Ricinus communis] |
| 20 |
Hb_132998_010 |
0.2644539121 |
- |
- |
hypothetical protein JCGZ_00197 [Jatropha curcas] |