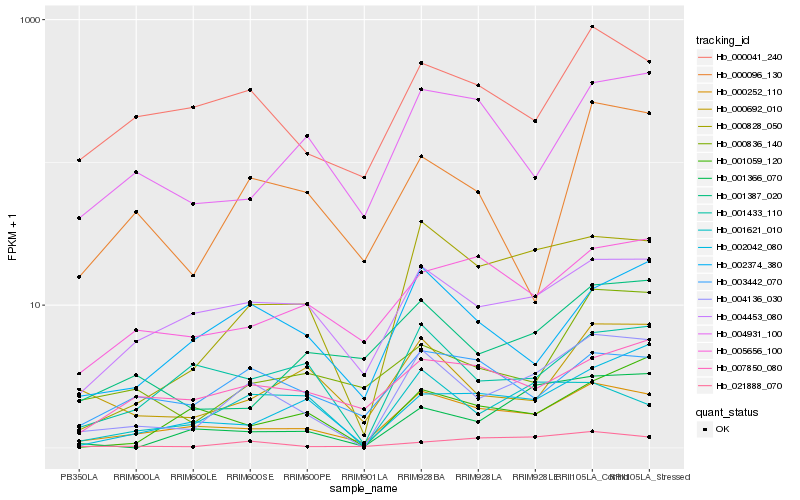
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004136_030 |
0.0 |
- |
- |
- |
| 2 |
Hb_000828_050 |
0.1690415707 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 3 |
Hb_000252_110 |
0.1721145192 |
- |
- |
- |
| 4 |
Hb_004453_080 |
0.2036339278 |
- |
- |
PREDICTED: acyl-protein thioesterase 2-like [Populus euphratica] |
| 5 |
Hb_001366_070 |
0.2100777247 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 4-like isoform X2 [Jatropha curcas] |
| 6 |
Hb_001433_110 |
0.2106029127 |
- |
- |
PREDICTED: uncharacterized protein LOC105647469 isoform X1 [Jatropha curcas] |
| 7 |
Hb_002374_380 |
0.2173870299 |
- |
- |
Acyl-CoA N-acyltransferases (NAT) superfamily protein isoform 1 [Theobroma cacao] |
| 8 |
Hb_021888_070 |
0.2213113213 |
- |
- |
Putative CCR4-associated factor 1 like 11 [Glycine soja] |
| 9 |
Hb_000692_010 |
0.2237388525 |
- |
- |
PREDICTED: uncharacterized protein LOC105635295 [Jatropha curcas] |
| 10 |
Hb_002042_080 |
0.2254626981 |
rubber biosynthesis |
Gene Name: Mevalonate kinase |
mevalonate kinase [Hevea brasiliensis] |
| 11 |
Hb_000041_240 |
0.227954018 |
- |
- |
glutaredoxin [Hevea brasiliensis] |
| 12 |
Hb_000836_140 |
0.2309206761 |
- |
- |
PREDICTED: protein TIC 20-IV, chloroplastic [Jatropha curcas] |
| 13 |
Hb_001387_020 |
0.2377286005 |
- |
- |
PREDICTED: uncharacterized protein LOC102587892 [Solanum tuberosum] |
| 14 |
Hb_001059_120 |
0.2391143199 |
- |
- |
NADH dehydrogenase, putative [Ricinus communis] |
| 15 |
Hb_001621_010 |
0.2400460255 |
- |
- |
PREDICTED: uncharacterized protein LOC102615432 [Citrus sinensis] |
| 16 |
Hb_000096_130 |
0.2469165646 |
- |
- |
hypothetical protein 23 [Hevea brasiliensis] |
| 17 |
Hb_007850_080 |
0.2478621617 |
- |
- |
PREDICTED: auxilin-like protein 1 isoform X1 [Jatropha curcas] |
| 18 |
Hb_005656_100 |
0.2487458859 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_003442_070 |
0.2514353986 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase SD1-8 [Jatropha curcas] |
| 20 |
Hb_004931_100 |
0.2532791332 |
- |
- |
Actin depolymerizing factor 6 isoform 1 [Theobroma cacao] |