| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
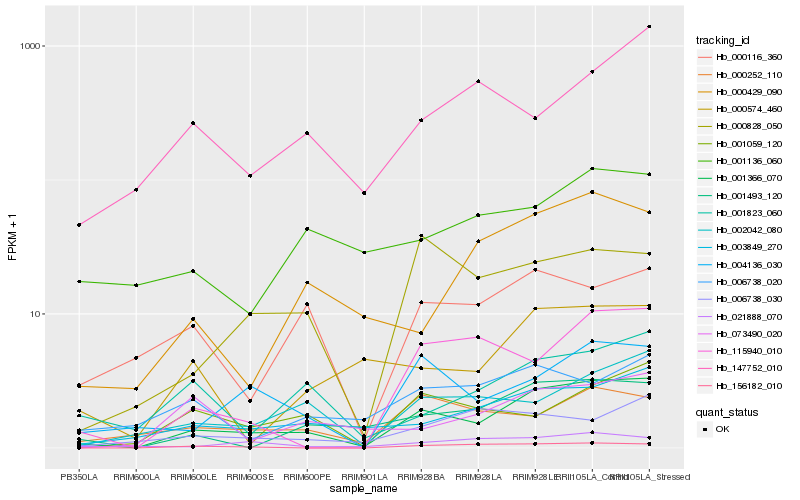
Hb_001366_070 |
0.0 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 4-like isoform X2 [Jatropha curcas] |
| 2 |
Hb_003849_270 |
0.2037984252 |
- |
- |
- |
| 3 |
Hb_001493_120 |
0.2095462964 |
- |
- |
- |
| 4 |
Hb_004136_030 |
0.2100777247 |
- |
- |
- |
| 5 |
Hb_002042_080 |
0.2115905724 |
rubber biosynthesis |
Gene Name: Mevalonate kinase |
mevalonate kinase [Hevea brasiliensis] |
| 6 |
Hb_000828_050 |
0.2260193017 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 7 |
Hb_001059_120 |
0.2278762205 |
- |
- |
NADH dehydrogenase, putative [Ricinus communis] |
| 8 |
Hb_156182_010 |
0.2309581853 |
- |
- |
PREDICTED: uncharacterized protein LOC105629587 [Jatropha curcas] |
| 9 |
Hb_115940_010 |
0.2367144596 |
- |
- |
putative auxin-amidohydrolase precursor family protein [Populus trichocarpa] |
| 10 |
Hb_001823_060 |
0.2376602376 |
- |
- |
fructose-bisphosphate aldolase, putative [Ricinus communis] |
| 11 |
Hb_000252_110 |
0.238218197 |
- |
- |
- |
| 12 |
Hb_000429_090 |
0.2387414809 |
- |
- |
PREDICTED: carboxyl-terminal-processing peptidase 3, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000574_460 |
0.2393518302 |
- |
- |
hypothetical protein JCGZ_06119 [Jatropha curcas] |
| 14 |
Hb_000116_360 |
0.2396802422 |
- |
- |
PREDICTED: uncharacterized protein LOC105116418 [Populus euphratica] |
| 15 |
Hb_021888_070 |
0.2462880665 |
- |
- |
Putative CCR4-associated factor 1 like 11 [Glycine soja] |
| 16 |
Hb_006738_030 |
0.2485671153 |
- |
- |
Ras-related protein Rab7 [Morus notabilis] |
| 17 |
Hb_073490_020 |
0.2502860479 |
- |
- |
hypothetical protein PRUPE_ppa016333mg, partial [Prunus persica] |
| 18 |
Hb_006738_020 |
0.2507933523 |
- |
- |
hypothetical protein VITISV_010510 [Vitis vinifera] |
| 19 |
Hb_001136_060 |
0.2580928945 |
- |
- |
PREDICTED: uncharacterized protein LOC105641994 [Jatropha curcas] |
| 20 |
Hb_147752_010 |
0.2602403821 |
- |
- |
PREDICTED: 40S ribosomal protein S9-2-like isoform X1 [Populus euphratica] |