| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
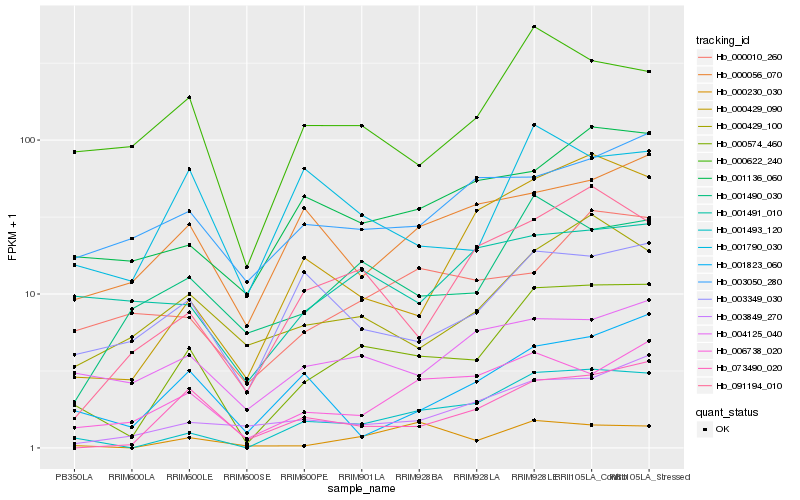
Hb_000574_460 |
0.0 |
- |
- |
hypothetical protein JCGZ_06119 [Jatropha curcas] |
| 2 |
Hb_001493_120 |
0.1418113944 |
- |
- |
- |
| 3 |
Hb_073490_020 |
0.1866758076 |
- |
- |
hypothetical protein PRUPE_ppa016333mg, partial [Prunus persica] |
| 4 |
Hb_001490_030 |
0.1916330025 |
- |
- |
PREDICTED: psbP domain-containing protein 6, chloroplastic [Jatropha curcas] |
| 5 |
Hb_091194_010 |
0.1925152241 |
- |
- |
hypothetical protein POPTR_0006s05430g [Populus trichocarpa] |
| 6 |
Hb_006738_020 |
0.1962421502 |
- |
- |
hypothetical protein VITISV_010510 [Vitis vinifera] |
| 7 |
Hb_000429_090 |
0.2004951773 |
- |
- |
PREDICTED: carboxyl-terminal-processing peptidase 3, chloroplastic [Jatropha curcas] |
| 8 |
Hb_003849_270 |
0.2007211346 |
- |
- |
- |
| 9 |
Hb_000429_100 |
0.2012282537 |
- |
- |
hypothetical protein POPTR_0006s05430g [Populus trichocarpa] |
| 10 |
Hb_000622_240 |
0.2022562865 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1A, chloroplastic [Jatropha curcas] |
| 11 |
Hb_001136_060 |
0.2058285774 |
- |
- |
PREDICTED: uncharacterized protein LOC105641994 [Jatropha curcas] |
| 12 |
Hb_000230_030 |
0.2074630424 |
- |
- |
hypothetical protein B456_013G2021001, partial [Gossypium raimondii] |
| 13 |
Hb_003349_030 |
0.2157966086 |
- |
- |
PREDICTED: uncharacterized protein LOC105628283 [Jatropha curcas] |
| 14 |
Hb_004125_040 |
0.2168078752 |
- |
- |
- |
| 15 |
Hb_000010_260 |
0.2182746355 |
- |
- |
PREDICTED: uncharacterized protein LOC105640273 [Jatropha curcas] |
| 16 |
Hb_001491_010 |
0.222052264 |
- |
- |
PREDICTED: uncharacterized protein LOC105649805 [Jatropha curcas] |
| 17 |
Hb_001790_030 |
0.2232983596 |
- |
- |
PREDICTED: ferredoxin-thioredoxin reductase catalytic chain, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000056_070 |
0.2243195207 |
- |
- |
- |
| 19 |
Hb_003050_280 |
0.2258109492 |
transcription factor |
TF Family: HMG |
DNA-binding protein MNB1B, putative [Ricinus communis] |
| 20 |
Hb_001823_060 |
0.2269465031 |
- |
- |
fructose-bisphosphate aldolase, putative [Ricinus communis] |