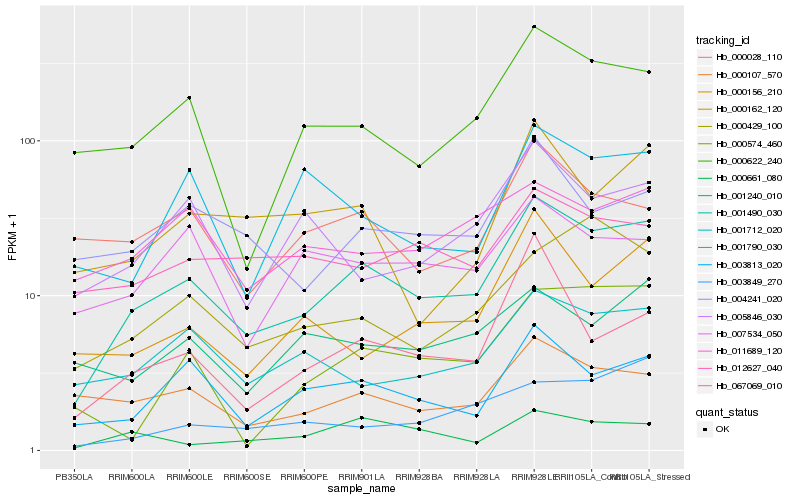
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001490_030 |
0.0 |
- |
- |
PREDICTED: psbP domain-containing protein 6, chloroplastic [Jatropha curcas] |
| 2 |
Hb_003813_020 |
0.1546803607 |
- |
- |
PREDICTED: uncharacterized protein LOC105638736 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000622_240 |
0.1587713551 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1A, chloroplastic [Jatropha curcas] |
| 4 |
Hb_001712_020 |
0.1703220496 |
- |
- |
PREDICTED: uncharacterized protein LOC105641948 [Jatropha curcas] |
| 5 |
Hb_000107_570 |
0.1770989002 |
- |
- |
PREDICTED: uncharacterized protein LOC105634044 isoform X1 [Jatropha curcas] |
| 6 |
Hb_004241_020 |
0.1775889126 |
- |
- |
PREDICTED: DAG protein, chloroplastic-like [Jatropha curcas] |
| 7 |
Hb_000156_210 |
0.1831821902 |
- |
- |
PREDICTED: metal transporter Nramp1 [Jatropha curcas] |
| 8 |
Hb_000429_100 |
0.1833124303 |
- |
- |
hypothetical protein POPTR_0006s05430g [Populus trichocarpa] |
| 9 |
Hb_005846_030 |
0.1843578937 |
- |
- |
PREDICTED: 50S ribosomal protein L29, chloroplastic [Jatropha curcas] |
| 10 |
Hb_067069_010 |
0.1844763568 |
- |
- |
hypothetical protein JCGZ_11524 [Jatropha curcas] |
| 11 |
Hb_011689_120 |
0.1854331974 |
- |
- |
PREDICTED: uncharacterized protein At4g15545 [Jatropha curcas] |
| 12 |
Hb_001240_010 |
0.1866740274 |
- |
- |
PREDICTED: uncharacterized protein LOC105634101 [Jatropha curcas] |
| 13 |
Hb_012627_040 |
0.1889233617 |
- |
- |
PREDICTED: uncharacterized protein LOC105641988 [Jatropha curcas] |
| 14 |
Hb_000162_120 |
0.1891911861 |
- |
- |
PREDICTED: chaperone protein dnaJ 11, chloroplastic-like [Jatropha curcas] |
| 15 |
Hb_000028_110 |
0.1894353961 |
- |
- |
PREDICTED: uncharacterized protein LOC105645305 [Jatropha curcas] |
| 16 |
Hb_003849_270 |
0.1906337747 |
- |
- |
- |
| 17 |
Hb_007534_050 |
0.1909537545 |
- |
- |
PREDICTED: glutamyl-tRNA reductase-binding protein, chloroplastic isoform X1 [Jatropha curcas] |
| 18 |
Hb_000661_080 |
0.1914877711 |
- |
- |
monoxygenase, putative [Ricinus communis] |
| 19 |
Hb_000574_460 |
0.1916330025 |
- |
- |
hypothetical protein JCGZ_06119 [Jatropha curcas] |
| 20 |
Hb_001790_030 |
0.1916814493 |
- |
- |
PREDICTED: ferredoxin-thioredoxin reductase catalytic chain, chloroplastic [Jatropha curcas] |