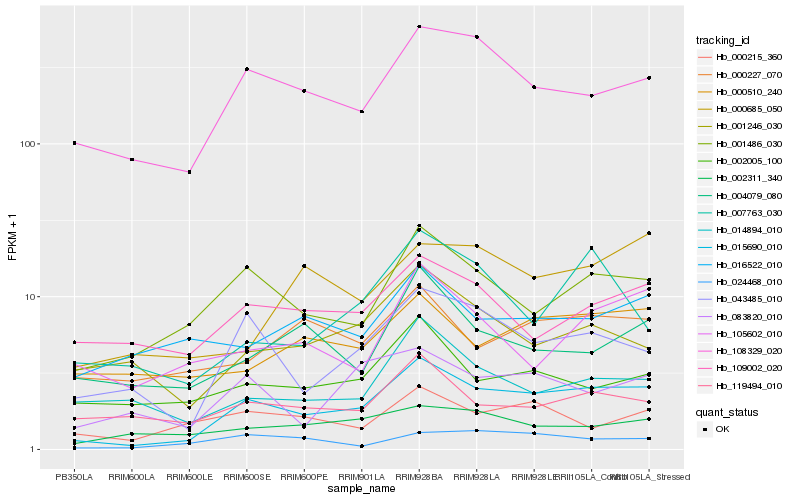
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_015690_010 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein CISIN_1g038521mg [Citrus sinensis] |
| 2 |
Hb_001486_030 |
0.162515088 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_108329_020 |
0.1677109987 |
transcription factor |
TF Family: EIL |
ETHYLENE-INSENSITIVE3 protein, putative [Ricinus communis] |
| 4 |
Hb_043485_010 |
0.168889635 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like isoform X2 [Jatropha curcas] |
| 5 |
Hb_083820_010 |
0.1956143772 |
- |
- |
hypothetical protein JCGZ_20198 [Jatropha curcas] |
| 6 |
Hb_000227_070 |
0.2016155573 |
- |
- |
PREDICTED: ATP-dependent (S)-NAD(P)H-hydrate dehydratase isoform X2 [Jatropha curcas] |
| 7 |
Hb_001246_030 |
0.201789028 |
- |
- |
hypothetical protein JCGZ_14672 [Jatropha curcas] |
| 8 |
Hb_109002_020 |
0.202932881 |
- |
- |
PREDICTED: CTP synthase-like [Jatropha curcas] |
| 9 |
Hb_000510_240 |
0.2035842126 |
desease resistance |
Gene Name: PRK |
ATP binding protein, putative [Ricinus communis] |
| 10 |
Hb_000685_050 |
0.2055269873 |
- |
- |
PREDICTED: mitochondrial inner membrane protein OXA1-like [Populus euphratica] |
| 11 |
Hb_119494_010 |
0.2058997201 |
- |
- |
PREDICTED: box C/D snoRNA protein 1 [Jatropha curcas] |
| 12 |
Hb_002311_340 |
0.2067198614 |
- |
- |
phospholipase d zeta, putative [Ricinus communis] |
| 13 |
Hb_024468_010 |
0.2069120776 |
- |
- |
hypothetical protein CISIN_1g046520mg [Citrus sinensis] |
| 14 |
Hb_105602_010 |
0.2071969784 |
- |
- |
Ketose-bisphosphate aldolase class-II family protein isoform 5 [Theobroma cacao] |
| 15 |
Hb_002005_100 |
0.207833614 |
- |
- |
- |
| 16 |
Hb_004079_080 |
0.207933036 |
- |
- |
PREDICTED: ras-related protein RHN1-like [Jatropha curcas] |
| 17 |
Hb_016522_010 |
0.2093918456 |
- |
- |
PREDICTED: short-chain dehydrogenase/reductase 2b-like [Jatropha curcas] |
| 18 |
Hb_014894_010 |
0.2097771849 |
- |
- |
hypothetical protein CICLE_v10001869mg [Citrus clementina] |
| 19 |
Hb_000215_360 |
0.2099043118 |
- |
- |
Uncharacterized protein isoform 2 [Theobroma cacao] |
| 20 |
Hb_007763_030 |
0.2115492821 |
- |
- |
PREDICTED: uncharacterized protein LOC105638154 [Jatropha curcas] |