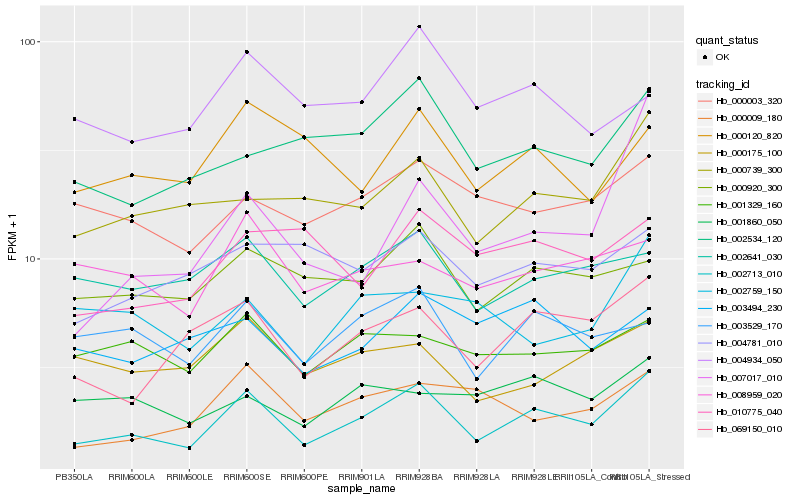
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002713_010 |
0.0 |
- |
- |
multidrug resistance protein 1, 2, putative [Ricinus communis] |
| 2 |
Hb_069150_010 |
0.1342987675 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPM1, putative [Ricinus communis] |
| 3 |
Hb_000739_300 |
0.1511694284 |
- |
- |
PREDICTED: uncharacterized protein LOC105643092 [Jatropha curcas] |
| 4 |
Hb_002534_120 |
0.1595228364 |
- |
- |
PREDICTED: protein DCL, chloroplastic [Jatropha curcas] |
| 5 |
Hb_003529_170 |
0.1613059829 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX29 [Jatropha curcas] |
| 6 |
Hb_000009_180 |
0.1626470625 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
RecName: Full=Geranylgeranyl pyrophosphate synthase, chloroplastic; Short=GGPP synthase; Short=GGPS; AltName: Full=(2E,6E)-farnesyl diphosphate synthase; AltName: Full=Dimethylallyltranstransferase; AltName: Full=Farnesyl diphosphate synthase; AltName: Full=Farnesyltranstransferase; AltName: Full=Geranyltranstransferase; Flags: Precursor [Hevea brasiliensis] |
| 7 |
Hb_000920_300 |
0.1634493673 |
- |
- |
PREDICTED: flowering time control protein FY isoform X2 [Jatropha curcas] |
| 8 |
Hb_002641_030 |
0.1660910654 |
- |
- |
PREDICTED: flowering time control protein FCA isoform X1 [Jatropha curcas] |
| 9 |
Hb_000003_320 |
0.1666327001 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 24 [Jatropha curcas] |
| 10 |
Hb_001329_160 |
0.1711486066 |
- |
- |
PREDICTED: uncharacterized protein LOC105642679 isoform X1 [Jatropha curcas] |
| 11 |
Hb_007017_010 |
0.1719307898 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor MYB1R1 [Jatropha curcas] |
| 12 |
Hb_000120_820 |
0.1731466124 |
- |
- |
dihydrodipicolinate synthase, putative [Ricinus communis] |
| 13 |
Hb_000175_100 |
0.1746020386 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 14 |
Hb_001860_050 |
0.1747345338 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g08070-like [Jatropha curcas] |
| 15 |
Hb_003494_230 |
0.1748049925 |
- |
- |
starch synthase isoform II [Manihot esculenta] |
| 16 |
Hb_004781_010 |
0.1753572993 |
- |
- |
PREDICTED: potassium transporter 11-like [Populus euphratica] |
| 17 |
Hb_002759_150 |
0.1763464156 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 22 isoform X1 [Jatropha curcas] |
| 18 |
Hb_008959_020 |
0.1764959403 |
- |
- |
PREDICTED: nitric oxide synthase-interacting protein [Pyrus x bretschneideri] |
| 19 |
Hb_010775_040 |
0.1767544107 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 4 [Jatropha curcas] |
| 20 |
Hb_004934_050 |
0.1773460591 |
- |
- |
Eukaryotic release factor 1-3 isoform 1 [Theobroma cacao] |