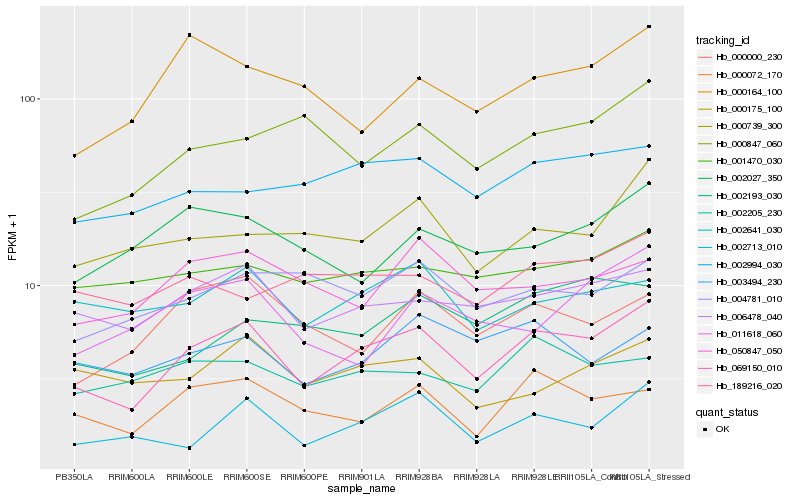
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_069150_010 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPM1, putative [Ricinus communis] |
| 2 |
Hb_006478_040 |
0.119584546 |
- |
- |
PREDICTED: methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial [Jatropha curcas] |
| 3 |
Hb_002713_010 |
0.1342987675 |
- |
- |
multidrug resistance protein 1, 2, putative [Ricinus communis] |
| 4 |
Hb_000072_170 |
0.1344313607 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FtsH [Jatropha curcas] |
| 5 |
Hb_002994_030 |
0.1358858403 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM22-like [Pyrus x bretschneideri] |
| 6 |
Hb_000739_300 |
0.1384652426 |
- |
- |
PREDICTED: uncharacterized protein LOC105643092 [Jatropha curcas] |
| 7 |
Hb_000000_230 |
0.1412124835 |
transcription factor |
TF Family: LIM |
PREDICTED: protein DA1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_002641_030 |
0.1421023666 |
- |
- |
PREDICTED: flowering time control protein FCA isoform X1 [Jatropha curcas] |
| 9 |
Hb_001470_030 |
0.1424446331 |
- |
- |
PREDICTED: MORC family CW-type zinc finger protein 3 isoform X2 [Jatropha curcas] |
| 10 |
Hb_003494_230 |
0.1431399334 |
- |
- |
starch synthase isoform II [Manihot esculenta] |
| 11 |
Hb_004781_010 |
0.1432497311 |
- |
- |
PREDICTED: potassium transporter 11-like [Populus euphratica] |
| 12 |
Hb_000847_060 |
0.1443239831 |
- |
- |
hypothetical protein [Jatropha curcas] |
| 13 |
Hb_002193_030 |
0.144779359 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH130-like [Jatropha curcas] |
| 14 |
Hb_050847_050 |
0.1461542009 |
- |
- |
PREDICTED: uncharacterized protein LOC105649140 isoform X1 [Jatropha curcas] |
| 15 |
Hb_002205_230 |
0.1473326753 |
- |
- |
glycerol-3-phosphate dehydrogenase, putative [Ricinus communis] |
| 16 |
Hb_000164_100 |
0.1511186728 |
- |
- |
PREDICTED: probable histone H2A.1 [Jatropha curcas] |
| 17 |
Hb_002027_350 |
0.151466119 |
- |
- |
PREDICTED: U1 small nuclear ribonucleoprotein C-like [Jatropha curcas] |
| 18 |
Hb_189216_020 |
0.1521251932 |
- |
- |
PREDICTED: translin [Jatropha curcas] |
| 19 |
Hb_011618_060 |
0.1535658817 |
- |
- |
PREDICTED: DNA-repair protein XRCC1 [Jatropha curcas] |
| 20 |
Hb_000175_100 |
0.154037331 |
- |
- |
protein kinase, putative [Ricinus communis] |