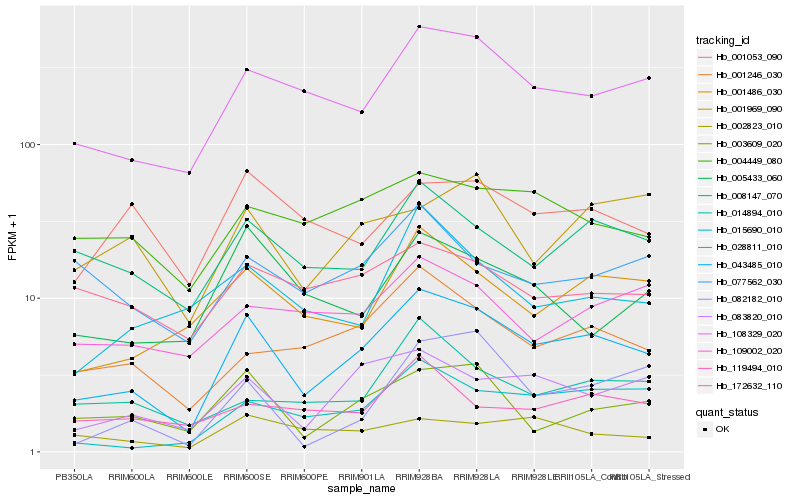
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_043485_010 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like isoform X2 [Jatropha curcas] |
| 2 |
Hb_108329_020 |
0.1418244084 |
transcription factor |
TF Family: EIL |
ETHYLENE-INSENSITIVE3 protein, putative [Ricinus communis] |
| 3 |
Hb_083820_010 |
0.159200528 |
- |
- |
hypothetical protein JCGZ_20198 [Jatropha curcas] |
| 4 |
Hb_001486_030 |
0.1658249486 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_015690_010 |
0.168889635 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein CISIN_1g038521mg [Citrus sinensis] |
| 6 |
Hb_001246_030 |
0.1690269683 |
- |
- |
hypothetical protein JCGZ_14672 [Jatropha curcas] |
| 7 |
Hb_008147_070 |
0.1714783335 |
- |
- |
PREDICTED: tryptophan synthase beta chain 1 [Jatropha curcas] |
| 8 |
Hb_003609_020 |
0.1752716484 |
desease resistance |
Gene Name: Clusterin |
hypothetical protein CISIN_1g001897mg [Citrus sinensis] |
| 9 |
Hb_004449_080 |
0.1878070548 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH3-like [Jatropha curcas] |
| 10 |
Hb_005433_060 |
0.1929621803 |
- |
- |
PREDICTED: putative 1-phosphatidylinositol-3-phosphate 5-kinase FAB1C [Jatropha curcas] |
| 11 |
Hb_028811_010 |
0.1936829745 |
- |
- |
PREDICTED: uncharacterized protein LOC105650178 isoform X1 [Jatropha curcas] |
| 12 |
Hb_014894_010 |
0.1953798061 |
- |
- |
hypothetical protein CICLE_v10001869mg [Citrus clementina] |
| 13 |
Hb_109002_020 |
0.1956738926 |
- |
- |
PREDICTED: CTP synthase-like [Jatropha curcas] |
| 14 |
Hb_001053_090 |
0.1968451565 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 30-like [Jatropha curcas] |
| 15 |
Hb_001969_090 |
0.1977581899 |
- |
- |
plastid ATP/ADP transport protein 2 [Manihot esculenta] |
| 16 |
Hb_077562_030 |
0.1992197158 |
- |
- |
PREDICTED: cytochrome P450 4X1-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_002823_010 |
0.1995724443 |
- |
- |
- |
| 18 |
Hb_172632_110 |
0.2008201903 |
- |
- |
PREDICTED: pumilio homolog 5 [Jatropha curcas] |
| 19 |
Hb_119494_010 |
0.2016221479 |
- |
- |
PREDICTED: box C/D snoRNA protein 1 [Jatropha curcas] |
| 20 |
Hb_082182_010 |
0.2033163731 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |