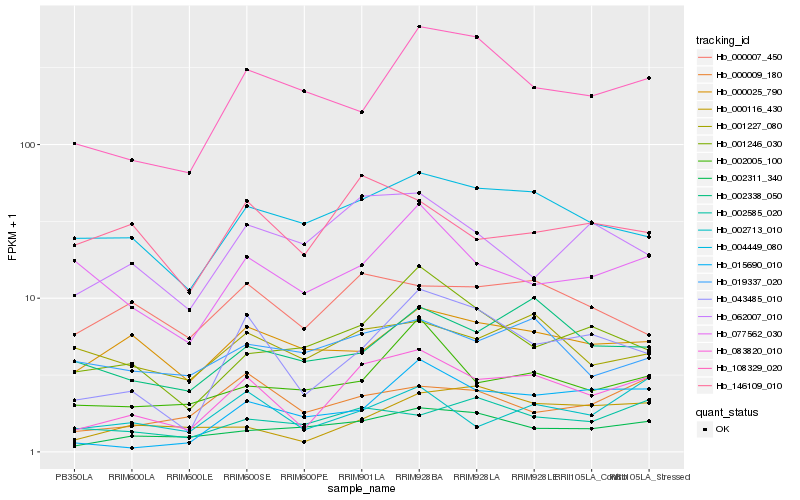
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_083820_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_20198 [Jatropha curcas] |
| 2 |
Hb_043485_010 |
0.159200528 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like isoform X2 [Jatropha curcas] |
| 3 |
Hb_002311_340 |
0.174356154 |
- |
- |
phospholipase d zeta, putative [Ricinus communis] |
| 4 |
Hb_004449_080 |
0.1745912408 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH3-like [Jatropha curcas] |
| 5 |
Hb_002005_100 |
0.1838279431 |
- |
- |
- |
| 6 |
Hb_002713_010 |
0.1843438997 |
- |
- |
multidrug resistance protein 1, 2, putative [Ricinus communis] |
| 7 |
Hb_000009_180 |
0.1890032551 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
RecName: Full=Geranylgeranyl pyrophosphate synthase, chloroplastic; Short=GGPP synthase; Short=GGPS; AltName: Full=(2E,6E)-farnesyl diphosphate synthase; AltName: Full=Dimethylallyltranstransferase; AltName: Full=Farnesyl diphosphate synthase; AltName: Full=Farnesyltranstransferase; AltName: Full=Geranyltranstransferase; Flags: Precursor [Hevea brasiliensis] |
| 8 |
Hb_062007_010 |
0.1911043868 |
- |
- |
PREDICTED: argininosuccinate lyase, chloroplastic [Jatropha curcas] |
| 9 |
Hb_002338_050 |
0.19247471 |
- |
- |
PREDICTED: uncharacterized protein LOC105118489 isoform X1 [Populus euphratica] |
| 10 |
Hb_146109_010 |
0.1942545055 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001227_080 |
0.1943949561 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 12 |
Hb_077562_030 |
0.1948970862 |
- |
- |
PREDICTED: cytochrome P450 4X1-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_015690_010 |
0.1956143772 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein CISIN_1g038521mg [Citrus sinensis] |
| 14 |
Hb_000007_450 |
0.1958871488 |
- |
- |
PREDICTED: tRNA(adenine(34)) deaminase, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000116_430 |
0.1966164558 |
- |
- |
PREDICTED: mitochondrial phosphate carrier protein 1, mitochondrial-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_001246_030 |
0.1967626488 |
- |
- |
hypothetical protein JCGZ_14672 [Jatropha curcas] |
| 17 |
Hb_019337_020 |
0.197677649 |
- |
- |
K+ uptake permease 11 isoform 1 [Theobroma cacao] |
| 18 |
Hb_000025_790 |
0.1985339938 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 3 [Jatropha curcas] |
| 19 |
Hb_108329_020 |
0.1990230357 |
transcription factor |
TF Family: EIL |
ETHYLENE-INSENSITIVE3 protein, putative [Ricinus communis] |
| 20 |
Hb_002585_020 |
0.199075166 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g18485-like [Populus euphratica] |