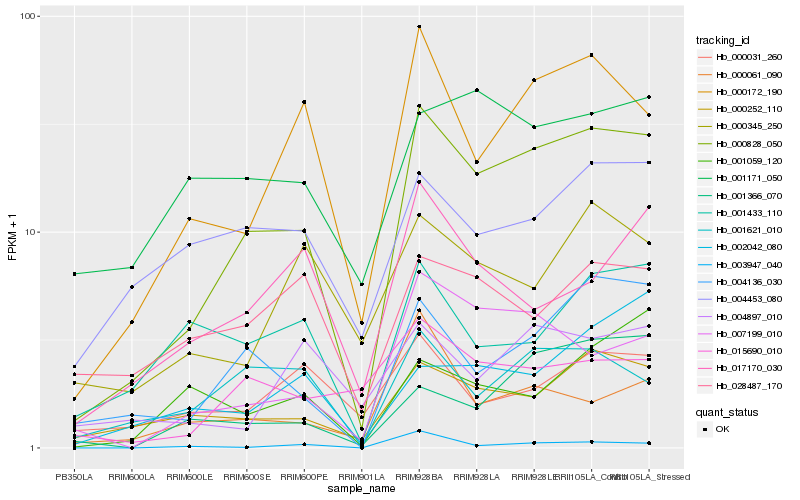
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000828_050 |
0.0 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 2 |
Hb_004136_030 |
0.1690415707 |
- |
- |
- |
| 3 |
Hb_000172_190 |
0.1692959165 |
- |
- |
PREDICTED: uncharacterized protein LOC105650824 [Jatropha curcas] |
| 4 |
Hb_000252_110 |
0.1751191763 |
- |
- |
- |
| 5 |
Hb_007199_010 |
0.1900900705 |
- |
- |
PREDICTED: protein kinase APK1A, chloroplastic [Nelumbo nucifera] |
| 6 |
Hb_001621_010 |
0.2041691246 |
- |
- |
PREDICTED: uncharacterized protein LOC102615432 [Citrus sinensis] |
| 7 |
Hb_015690_010 |
0.2156104112 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein CISIN_1g038521mg [Citrus sinensis] |
| 8 |
Hb_001171_050 |
0.2198321042 |
- |
- |
PREDICTED: uncharacterized protein DDB_G0284459 [Jatropha curcas] |
| 9 |
Hb_004453_080 |
0.220154762 |
- |
- |
PREDICTED: acyl-protein thioesterase 2-like [Populus euphratica] |
| 10 |
Hb_017170_030 |
0.2226718512 |
- |
- |
PREDICTED: acyl-coenzyme A oxidase 3, peroxisomal-like [Jatropha curcas] |
| 11 |
Hb_001433_110 |
0.2246881816 |
- |
- |
PREDICTED: uncharacterized protein LOC105647469 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000345_250 |
0.2252251502 |
- |
- |
cullin 1-like protein [Hevea brasiliensis] |
| 13 |
Hb_001366_070 |
0.2260193017 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 4-like isoform X2 [Jatropha curcas] |
| 14 |
Hb_000061_090 |
0.226723708 |
- |
- |
hypothetical protein B456_003G0981001, partial [Gossypium raimondii] |
| 15 |
Hb_002042_080 |
0.2309583475 |
rubber biosynthesis |
Gene Name: Mevalonate kinase |
mevalonate kinase [Hevea brasiliensis] |
| 16 |
Hb_028487_170 |
0.2334557109 |
- |
- |
PREDICTED: uncharacterized protein LOC105634088 [Jatropha curcas] |
| 17 |
Hb_004897_010 |
0.2338870862 |
- |
- |
- |
| 18 |
Hb_000031_260 |
0.2351280531 |
- |
- |
hypothetical protein POPTR_0018s03360g [Populus trichocarpa] |
| 19 |
Hb_001059_120 |
0.2373409553 |
- |
- |
NADH dehydrogenase, putative [Ricinus communis] |
| 20 |
Hb_003947_040 |
0.2386394332 |
- |
- |
Leucine-rich repeat transmembrane protein kinase protein, putative [Theobroma cacao] |