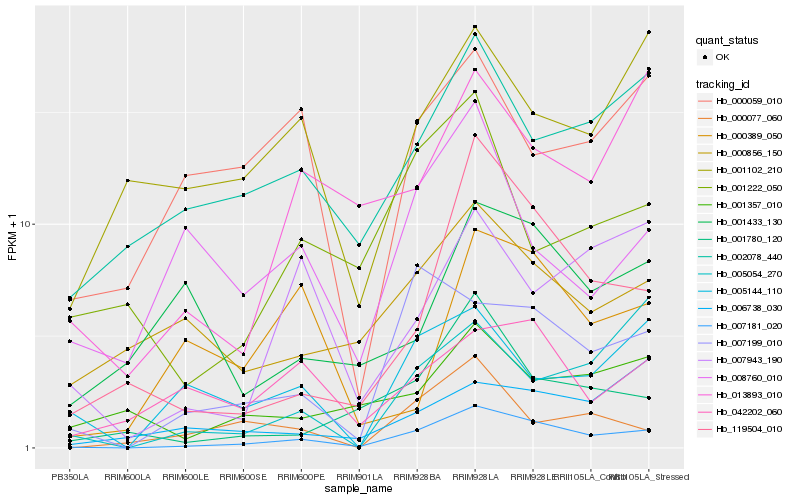
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007181_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein At4g04980-like [Populus euphratica] |
| 2 |
Hb_007199_010 |
0.22197766 |
- |
- |
PREDICTED: protein kinase APK1A, chloroplastic [Nelumbo nucifera] |
| 3 |
Hb_119504_010 |
0.2227912544 |
- |
- |
hypothetical protein JCGZ_15304 [Jatropha curcas] |
| 4 |
Hb_001780_120 |
0.2312854385 |
- |
- |
PREDICTED: uncharacterized protein LOC105634429 [Jatropha curcas] |
| 5 |
Hb_002078_440 |
0.2364142428 |
transcription factor |
TF Family: Orphans |
PREDICTED: histidine kinase 3 isoform X1 [Jatropha curcas] |
| 6 |
Hb_013893_010 |
0.2488609225 |
- |
- |
neutral/alkaline invertase 3 [Hevea brasiliensis] |
| 7 |
Hb_000856_150 |
0.2496616629 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g18900 [Jatropha curcas] |
| 8 |
Hb_042202_060 |
0.2550891675 |
- |
- |
PREDICTED: probable glycosyltransferase At5g03795 [Jatropha curcas] |
| 9 |
Hb_007943_190 |
0.2576502339 |
- |
- |
cytoplasmic dynein light chain, putative [Ricinus communis] |
| 10 |
Hb_005144_110 |
0.2584105217 |
- |
- |
PREDICTED: uncharacterized protein LOC104455487 [Eucalyptus grandis] |
| 11 |
Hb_005054_270 |
0.2605236254 |
- |
- |
- |
| 12 |
Hb_001222_050 |
0.2629409587 |
- |
- |
hypothetical protein CICLE_v100161911mg, partial [Citrus clementina] |
| 13 |
Hb_000077_060 |
0.2640087133 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH36-like [Jatropha curcas] |
| 14 |
Hb_001357_010 |
0.2643128533 |
- |
- |
- |
| 15 |
Hb_001102_210 |
0.2647023245 |
- |
- |
PREDICTED: alpha,alpha-trehalose-phosphate synthase [UDP-forming] 1-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_006738_030 |
0.2665968166 |
- |
- |
Ras-related protein Rab7 [Morus notabilis] |
| 17 |
Hb_000389_050 |
0.2692001419 |
- |
- |
PREDICTED: uncharacterized protein LOC105650917 [Jatropha curcas] |
| 18 |
Hb_000059_010 |
0.2706952664 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |
| 19 |
Hb_008760_010 |
0.2714241801 |
- |
- |
berberine bridge enzyme [Hevea brasiliensis] |
| 20 |
Hb_001433_130 |
0.2715611762 |
- |
- |
PREDICTED: uncharacterized protein LOC105647468 isoform X1 [Jatropha curcas] |