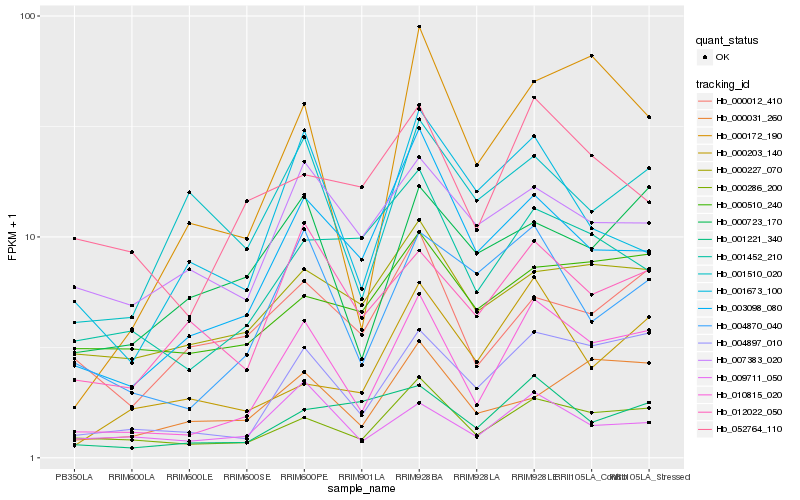
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010815_020 |
0.0 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 2 |
Hb_004897_010 |
0.1103038022 |
- |
- |
- |
| 3 |
Hb_000286_200 |
0.1426457864 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X2 [Jatropha curcas] |
| 4 |
Hb_004870_040 |
0.1693948045 |
- |
- |
- |
| 5 |
Hb_003098_080 |
0.1754675348 |
- |
- |
- |
| 6 |
Hb_000172_190 |
0.1793337281 |
- |
- |
PREDICTED: uncharacterized protein LOC105650824 [Jatropha curcas] |
| 7 |
Hb_012022_050 |
0.1810497145 |
- |
- |
hypothetical protein EUGRSUZ_B02804 [Eucalyptus grandis] |
| 8 |
Hb_000723_170 |
0.1904229751 |
- |
- |
PREDICTED: OTU domain-containing protein DDB_G0284757 [Jatropha curcas] |
| 9 |
Hb_000031_260 |
0.1908386796 |
- |
- |
hypothetical protein POPTR_0018s03360g [Populus trichocarpa] |
| 10 |
Hb_009711_050 |
0.1923022536 |
- |
- |
hypothetical protein POPTR_0019s04500g [Populus trichocarpa] |
| 11 |
Hb_001221_340 |
0.1946823877 |
- |
- |
hypothetical protein EUGRSUZ_B00102 [Eucalyptus grandis] |
| 12 |
Hb_001452_210 |
0.1967272209 |
- |
- |
molybdopterin biosynthesis CNX1 protein [Arabidopsis thaliana] |
| 13 |
Hb_000227_070 |
0.1994350934 |
- |
- |
PREDICTED: ATP-dependent (S)-NAD(P)H-hydrate dehydratase isoform X2 [Jatropha curcas] |
| 14 |
Hb_000012_410 |
0.2005995838 |
- |
- |
PREDICTED: histone deacetylase 2 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000203_140 |
0.2011718895 |
- |
- |
PREDICTED: peroxisome biogenesis protein 7 [Jatropha curcas] |
| 16 |
Hb_052764_110 |
0.2048129397 |
- |
- |
PREDICTED: quinone oxidoreductase PIG3 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001673_100 |
0.2067402591 |
- |
- |
PREDICTED: putative phosphatidylglycerol/phosphatidylinositol transfer protein DDB_G0282179 [Jatropha curcas] |
| 18 |
Hb_001510_020 |
0.2091111924 |
- |
- |
Diaminopimelate epimerase, putative [Ricinus communis] |
| 19 |
Hb_000510_240 |
0.2097338315 |
desease resistance |
Gene Name: PRK |
ATP binding protein, putative [Ricinus communis] |
| 20 |
Hb_007383_020 |
0.2101653711 |
- |
- |
conserved hypothetical protein [Ricinus communis] |