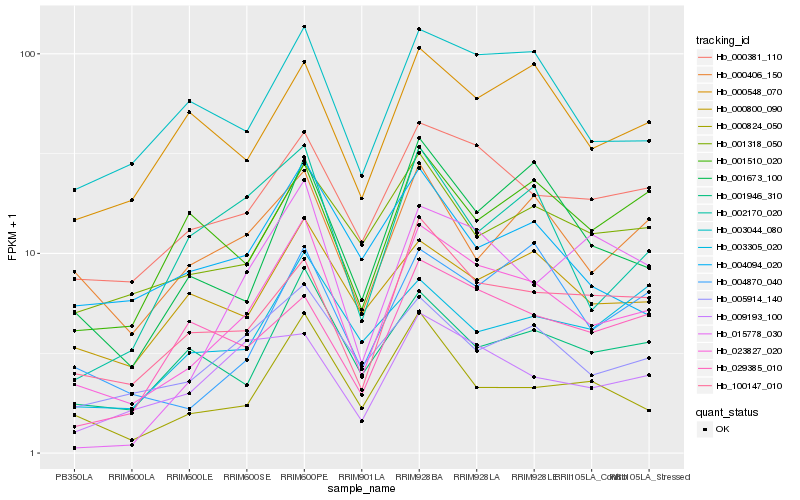
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_023827_020 |
0.0 |
- |
- |
beta-1,3-glucanase 1 [Ziziphus jujuba] |
| 2 |
Hb_000381_110 |
0.1243938667 |
- |
- |
PREDICTED: dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex 2, mitochondrial-like isoform X2 [Jatropha curcas] |
| 3 |
Hb_100147_010 |
0.1347972404 |
- |
- |
PREDICTED: exosome complex component MTR3 [Jatropha curcas] |
| 4 |
Hb_001673_100 |
0.1440675645 |
- |
- |
PREDICTED: putative phosphatidylglycerol/phosphatidylinositol transfer protein DDB_G0282179 [Jatropha curcas] |
| 5 |
Hb_000824_050 |
0.1446581713 |
- |
- |
PREDICTED: nicotinate phosphoribosyltransferase 1 [Jatropha curcas] |
| 6 |
Hb_005914_140 |
0.1469149137 |
- |
- |
PREDICTED: uncharacterized protein LOC105632183 [Jatropha curcas] |
| 7 |
Hb_003044_080 |
0.1496288832 |
desease resistance |
Gene Name: Synthase_beta |
RecName: Full=ATP synthase subunit beta, mitochondrial; Flags: Precursor [Hevea brasiliensis] |
| 8 |
Hb_001946_310 |
0.1519628335 |
- |
- |
PREDICTED: probable ethanolamine kinase isoform X2 [Jatropha curcas] |
| 9 |
Hb_004870_040 |
0.1563463205 |
- |
- |
- |
| 10 |
Hb_009193_100 |
0.1576418986 |
- |
- |
hypothetical protein JCGZ_15709 [Jatropha curcas] |
| 11 |
Hb_001318_050 |
0.158630164 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 12 |
Hb_002170_020 |
0.1614549527 |
transcription factor |
TF Family: MYB |
MYB transcription factor [Hevea brasiliensis] |
| 13 |
Hb_000800_090 |
0.163686759 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 1 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 1 of pyruvate dehydrogenase complex, mitochondrial [Jatropha curcas] |
| 14 |
Hb_000406_150 |
0.1642844583 |
- |
- |
sodium/hydrogen exchanger, putative [Ricinus communis] |
| 15 |
Hb_000548_070 |
0.1682910129 |
desease resistance |
Gene Name: Synthase_beta |
RecName: Full=ATP synthase subunit beta, mitochondrial; Flags: Precursor [Hevea brasiliensis] |
| 16 |
Hb_004094_020 |
0.173110911 |
- |
- |
Pyruvate kinase family protein isoform 1 [Theobroma cacao] |
| 17 |
Hb_029385_010 |
0.173428666 |
- |
- |
CTP synthase family protein [Populus trichocarpa] |
| 18 |
Hb_015778_030 |
0.175175755 |
- |
- |
PREDICTED: probable myosin light chain kinase DDB_G0279831 [Jatropha curcas] |
| 19 |
Hb_001510_020 |
0.1753566606 |
- |
- |
Diaminopimelate epimerase, putative [Ricinus communis] |
| 20 |
Hb_003305_020 |
0.1757341729 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 13-like [Jatropha curcas] |