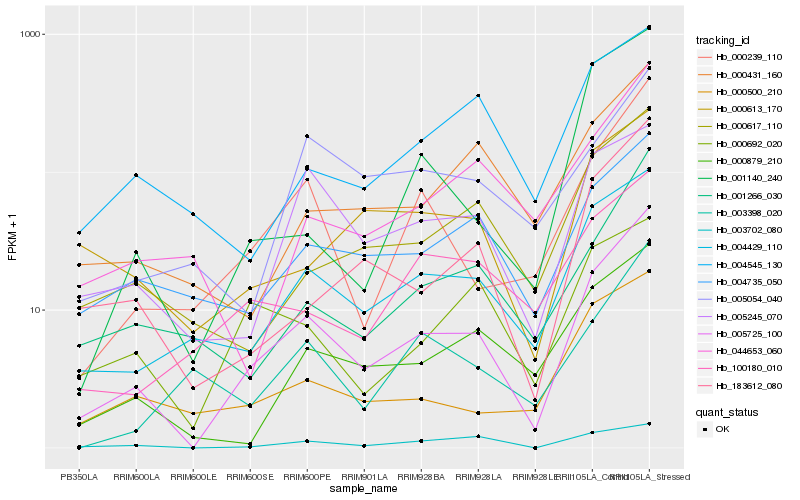
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005725_100 |
0.0 |
- |
- |
hypothetical protein 30 [Hevea brasiliensis] |
| 2 |
Hb_004429_110 |
0.1589987752 |
- |
- |
PREDICTED: protein SPIRAL1-like 1 [Populus euphratica] |
| 3 |
Hb_000239_110 |
0.1679843974 |
- |
- |
PREDICTED: adenine phosphoribosyltransferase 1-like [Jatropha curcas] |
| 4 |
Hb_183612_080 |
0.1736503083 |
- |
- |
hypothetical protein POPTR_0001s00500g [Populus trichocarpa] |
| 5 |
Hb_001266_030 |
0.1824477406 |
- |
- |
PREDICTED: lipid phosphate phosphatase epsilon 2, chloroplastic-like [Jatropha curcas] |
| 6 |
Hb_000879_210 |
0.1875653633 |
- |
- |
hypothetical protein POPTR_0014s01620g [Populus trichocarpa] |
| 7 |
Hb_000617_110 |
0.1884173174 |
- |
- |
translationally controlled tumor protein [Hevea brasiliensis] |
| 8 |
Hb_004735_050 |
0.1909905235 |
- |
- |
PREDICTED: uncharacterized protein LOC105639312 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000500_210 |
0.1944043157 |
- |
- |
- |
| 10 |
Hb_000431_160 |
0.1996167421 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit gamma-like [Jatropha curcas] |
| 11 |
Hb_003702_080 |
0.2015213181 |
- |
- |
cysteine protease, putative [Ricinus communis] |
| 12 |
Hb_100180_010 |
0.2063618071 |
- |
- |
PREDICTED: disease resistance protein RPM1-like [Jatropha curcas] |
| 13 |
Hb_003398_020 |
0.206871867 |
- |
- |
PREDICTED: uncharacterized protein LOC105647439 [Jatropha curcas] |
| 14 |
Hb_000613_170 |
0.2073065759 |
- |
- |
PREDICTED: RPM1-interacting protein 4 [Jatropha curcas] |
| 15 |
Hb_044653_060 |
0.2084368646 |
- |
- |
PREDICTED: uncharacterized protein LOC105628750 [Jatropha curcas] |
| 16 |
Hb_005245_070 |
0.2100504864 |
- |
- |
PREDICTED: uncharacterized protein LOC105643658 [Jatropha curcas] |
| 17 |
Hb_001140_240 |
0.211213681 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 18 |
Hb_005054_040 |
0.2139433406 |
- |
- |
PREDICTED: 60S ribosomal protein L35a-1 [Gossypium raimondii] |
| 19 |
Hb_004545_130 |
0.2149316286 |
- |
- |
uncharacterized protein LOC100305465 [Glycine max] |
| 20 |
Hb_000692_020 |
0.2188595122 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase ATL23 [Jatropha curcas] |