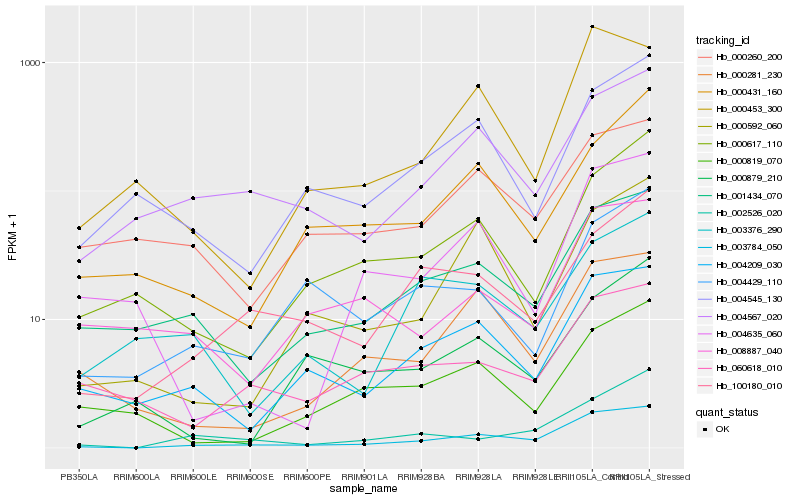
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003784_050 |
0.0 |
- |
- |
PREDICTED: replication protein A 32 kDa subunit B [Jatropha curcas] |
| 2 |
Hb_004209_030 |
0.1612711741 |
- |
- |
PREDICTED: (DL)-glycerol-3-phosphatase 2 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001434_070 |
0.1644697465 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM8 [Jatropha curcas] |
| 4 |
Hb_004567_020 |
0.1694571876 |
- |
- |
Small ubiquitin-related modifier 2 [Morus notabilis] |
| 5 |
Hb_002526_020 |
0.1747188309 |
- |
- |
- |
| 6 |
Hb_100180_010 |
0.1753945759 |
- |
- |
PREDICTED: disease resistance protein RPM1-like [Jatropha curcas] |
| 7 |
Hb_000281_230 |
0.1760643688 |
- |
- |
PREDICTED: probable lysine--tRNA ligase, cytoplasmic isoform X2 [Jatropha curcas] |
| 8 |
Hb_000617_110 |
0.187649626 |
- |
- |
translationally controlled tumor protein [Hevea brasiliensis] |
| 9 |
Hb_000592_060 |
0.1887598253 |
- |
- |
PREDICTED: ras-related protein RABA6a [Populus euphratica] |
| 10 |
Hb_004545_130 |
0.1903879853 |
- |
- |
uncharacterized protein LOC100305465 [Glycine max] |
| 11 |
Hb_000453_300 |
0.191283275 |
- |
- |
latex cystatin [Hevea brasiliensis] |
| 12 |
Hb_060618_010 |
0.1930896519 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 13 |
Hb_004429_110 |
0.1959133948 |
- |
- |
PREDICTED: protein SPIRAL1-like 1 [Populus euphratica] |
| 14 |
Hb_000431_160 |
0.1982762959 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit gamma-like [Jatropha curcas] |
| 15 |
Hb_000260_200 |
0.2006266708 |
- |
- |
- |
| 16 |
Hb_008887_040 |
0.2025177227 |
- |
- |
Ribonuclease P family protein / Rpp14 family protein isoform 1 [Theobroma cacao] |
| 17 |
Hb_000819_070 |
0.2106660017 |
- |
- |
PREDICTED: dolichol-phosphate mannosyltransferase subunit 1 [Jatropha curcas] |
| 18 |
Hb_000879_210 |
0.2119773192 |
- |
- |
hypothetical protein POPTR_0014s01620g [Populus trichocarpa] |
| 19 |
Hb_003376_290 |
0.2121064151 |
- |
- |
hypothetical protein POPTR_0010s22880g [Populus trichocarpa] |
| 20 |
Hb_004635_060 |
0.2135449842 |
- |
- |
hypothetical protein POPTR_0006s22850g [Populus trichocarpa] |