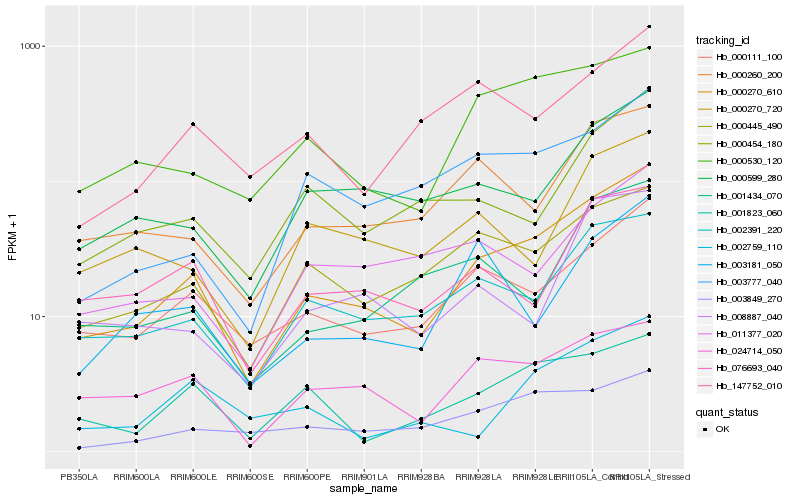
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000270_610 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein At5g50100, mitochondrial [Jatropha curcas] |
| 2 |
Hb_000111_100 |
0.138898755 |
- |
- |
PREDICTED: NADPH:quinone oxidoreductase-like [Camelina sativa] |
| 3 |
Hb_002391_220 |
0.1468925022 |
- |
- |
PREDICTED: UPF0548 protein At2g17695 [Jatropha curcas] |
| 4 |
Hb_000530_120 |
0.1583960833 |
- |
- |
copper transport protein ATOX1 [Hevea brasiliensis] |
| 5 |
Hb_000445_490 |
0.1612305307 |
- |
- |
- |
| 6 |
Hb_000260_200 |
0.1617473211 |
- |
- |
- |
| 7 |
Hb_000599_280 |
0.1635122427 |
- |
- |
PREDICTED: 60S ribosomal protein L14-1 [Jatropha curcas] |
| 8 |
Hb_001434_070 |
0.1718435866 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM8 [Jatropha curcas] |
| 9 |
Hb_076693_040 |
0.1740008602 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 5-B [Tarenaya hassleriana] |
| 10 |
Hb_000454_180 |
0.174039683 |
- |
- |
PREDICTED: uncharacterized protein LOC105134496 [Populus euphratica] |
| 11 |
Hb_003849_270 |
0.1744117952 |
- |
- |
- |
| 12 |
Hb_024714_050 |
0.1749519408 |
- |
- |
hypothetical protein CICLE_v100188992mg, partial [Citrus clementina] |
| 13 |
Hb_001823_060 |
0.1754717721 |
- |
- |
fructose-bisphosphate aldolase, putative [Ricinus communis] |
| 14 |
Hb_003777_040 |
0.1772023382 |
- |
- |
PREDICTED: sm-like protein LSM3A [Jatropha curcas] |
| 15 |
Hb_147752_010 |
0.1803129431 |
- |
- |
PREDICTED: 40S ribosomal protein S9-2-like isoform X1 [Populus euphratica] |
| 16 |
Hb_008887_040 |
0.1804500168 |
- |
- |
Ribonuclease P family protein / Rpp14 family protein isoform 1 [Theobroma cacao] |
| 17 |
Hb_011377_020 |
0.1809730222 |
- |
- |
hypothetical protein CARUB_v10018269mg, partial [Capsella rubella] |
| 18 |
Hb_000270_720 |
0.1813030356 |
- |
- |
hypothetical protein POPTR_0015s08460g [Populus trichocarpa] |
| 19 |
Hb_003181_050 |
0.1873098272 |
- |
- |
PREDICTED: DNA replication complex GINS protein PSF2 [Jatropha curcas] |
| 20 |
Hb_002759_110 |
0.1885752474 |
- |
- |
RNA-binding family protein, putative isoform 1 [Theobroma cacao] |