| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
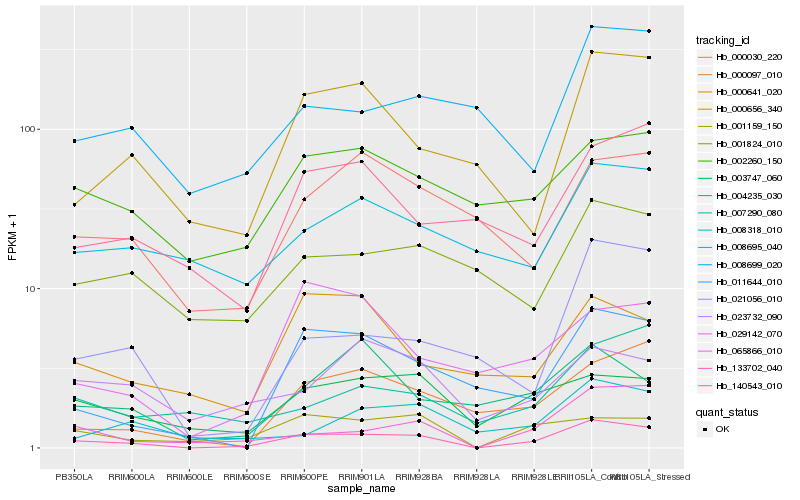
Hb_133702_040 |
0.0 |
- |
- |
- |
| 2 |
Hb_065866_010 |
0.2097940473 |
- |
- |
PREDICTED: hydroxyacyl-thioester dehydratase type 2, mitochondrial [Fragaria vesca subsp. vesca] |
| 3 |
Hb_001159_150 |
0.2213387001 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 2, chloroplastic isoform X2 [Jatropha curcas] |
| 4 |
Hb_011644_010 |
0.227320782 |
- |
- |
PREDICTED: probable choline-phosphate cytidylyltransferase [Elaeis guineensis] |
| 5 |
Hb_003747_060 |
0.2326600741 |
- |
- |
PREDICTED: CMP-N-acetylneuraminate-beta-galactosamide-alpha-2,3-sialyltransferase 2 isoform X2 [Cucumis sativus] |
| 6 |
Hb_000097_010 |
0.2365720377 |
- |
- |
- |
| 7 |
Hb_000656_340 |
0.2391861059 |
- |
- |
hypothetical protein JCGZ_24686 [Jatropha curcas] |
| 8 |
Hb_021056_010 |
0.2409767531 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 9 |
Hb_002260_150 |
0.2506757112 |
- |
- |
PREDICTED: uncharacterized protein LOC105647187 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000030_220 |
0.2538048911 |
- |
- |
PREDICTED: uncharacterized protein LOC105650405 [Jatropha curcas] |
| 11 |
Hb_008318_010 |
0.2568599859 |
- |
- |
PREDICTED: WD repeat-containing protein 82-B isoform X1 [Jatropha curcas] |
| 12 |
Hb_140543_010 |
0.2571575896 |
- |
- |
- |
| 13 |
Hb_007290_080 |
0.2573911589 |
- |
- |
hypothetical protein CISIN_1g047098mg [Citrus sinensis] |
| 14 |
Hb_023732_090 |
0.2580998767 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 15 |
Hb_029142_070 |
0.2588700579 |
- |
- |
- |
| 16 |
Hb_000641_020 |
0.2601909695 |
- |
- |
PREDICTED: tubulin-folding cofactor B-like [Jatropha curcas] |
| 17 |
Hb_008695_040 |
0.2603415681 |
- |
- |
PREDICTED: small ribosomal subunit protein S13, mitochondrial-like [Jatropha curcas] |
| 18 |
Hb_004235_030 |
0.2604120538 |
- |
- |
alpha-2,8-sialyltransferase 8b, putative [Ricinus communis] |
| 19 |
Hb_008699_020 |
0.2604681936 |
- |
- |
eukaryotic translation initiation factor 5A isoform IV [Hevea brasiliensis] |
| 20 |
Hb_001824_010 |
0.2612069554 |
- |
- |
glutathione-s-transferase theta, gst, putative [Ricinus communis] |