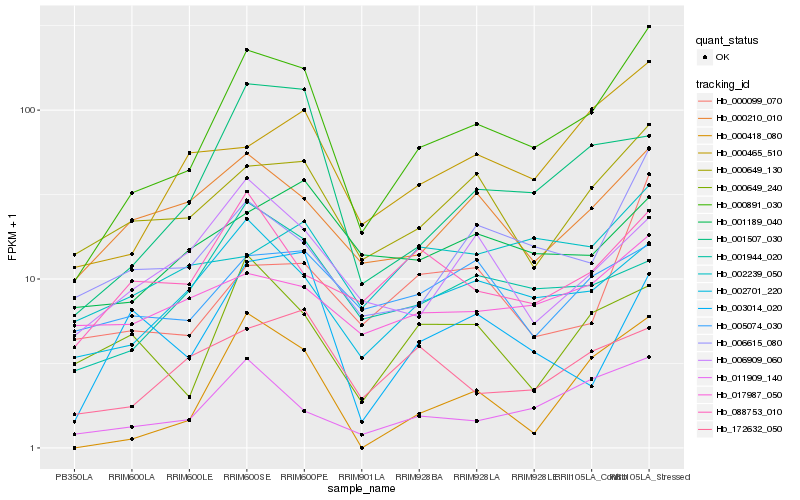
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000891_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein At4g22758 [Jatropha curcas] |
| 2 |
Hb_002701_220 |
0.1726931778 |
- |
- |
PREDICTED: solute carrier family 25 member 44 [Jatropha curcas] |
| 3 |
Hb_011909_140 |
0.1728748172 |
- |
- |
PREDICTED: diacylglycerol kinase 5-like [Jatropha curcas] |
| 4 |
Hb_000649_130 |
0.1741640899 |
- |
- |
Polcalcin Jun o, putative [Ricinus communis] |
| 5 |
Hb_000465_510 |
0.178717393 |
- |
- |
PREDICTED: GTP-binding protein YPTM2 [Populus euphratica] |
| 6 |
Hb_006615_080 |
0.1795259472 |
- |
- |
PREDICTED: uncharacterized protein LOC105640684 isoform X2 [Jatropha curcas] |
| 7 |
Hb_001507_030 |
0.1910708632 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 2 [Jatropha curcas] |
| 8 |
Hb_000210_010 |
0.1948256058 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 11 [Jatropha curcas] |
| 9 |
Hb_000418_080 |
0.1981244532 |
- |
- |
PREDICTED: uncharacterized protein LOC105124652 [Populus euphratica] |
| 10 |
Hb_001944_020 |
0.2049622616 |
- |
- |
PREDICTED: uncharacterized protein LOC105636535 [Jatropha curcas] |
| 11 |
Hb_000099_070 |
0.2086855236 |
- |
- |
PREDICTED: uncharacterized protein At5g41620 [Jatropha curcas] |
| 12 |
Hb_172632_050 |
0.2093159988 |
- |
- |
PREDICTED: uncharacterized protein LOC105646134 [Jatropha curcas] |
| 13 |
Hb_088753_010 |
0.2103499024 |
- |
- |
PREDICTED: copper amine oxidase 1-like [Vitis vinifera] |
| 14 |
Hb_006909_060 |
0.2131950977 |
- |
- |
PREDICTED: eukaryotic peptide chain release factor subunit 1-3-like [Solanum lycopersicum] |
| 15 |
Hb_002239_050 |
0.214476369 |
- |
- |
PREDICTED: isovaleryl-CoA dehydrogenase, mitochondrial [Jatropha curcas] |
| 16 |
Hb_000649_240 |
0.214662985 |
- |
- |
PREDICTED: probable peroxygenase 4 [Jatropha curcas] |
| 17 |
Hb_003014_020 |
0.2153943089 |
- |
- |
electron transporter, putative [Ricinus communis] |
| 18 |
Hb_001189_040 |
0.2156739295 |
- |
- |
hypothetical protein POPTR_0014s10420g [Populus trichocarpa] |
| 19 |
Hb_017987_050 |
0.2159317289 |
- |
- |
PREDICTED: branched-chain-amino-acid aminotransferase 5, chloroplastic-like [Jatropha curcas] |
| 20 |
Hb_005074_030 |
0.2182296094 |
- |
- |
- |