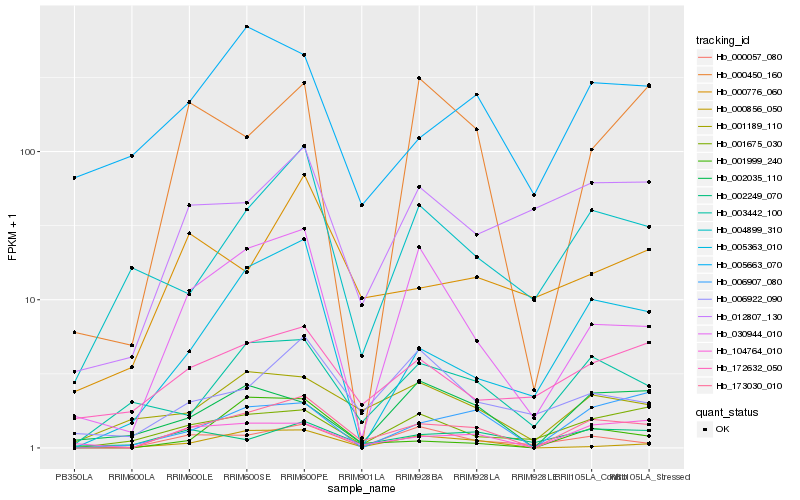
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_173030_010 |
0.0 |
- |
- |
hypothetical protein RCOM_1321060 [Ricinus communis] |
| 2 |
Hb_104764_010 |
0.2070024713 |
- |
- |
hypothetical protein JCGZ_16024 [Jatropha curcas] |
| 3 |
Hb_000057_080 |
0.2284569245 |
- |
- |
PREDICTED: probable ATP-dependent RNA helicase DDX11 [Jatropha curcas] |
| 4 |
Hb_005363_010 |
0.2288685782 |
- |
- |
hypothetical protein JCGZ_13835 [Jatropha curcas] |
| 5 |
Hb_030944_010 |
0.2318097163 |
- |
- |
PREDICTED: uncharacterized protein LOC105633455 [Jatropha curcas] |
| 6 |
Hb_003442_100 |
0.2321582898 |
- |
- |
PREDICTED: uncharacterized protein LOC105650128 [Jatropha curcas] |
| 7 |
Hb_006907_080 |
0.2398545884 |
- |
- |
PREDICTED: RPM1-interacting protein 4-like [Jatropha curcas] |
| 8 |
Hb_001189_110 |
0.2405112009 |
- |
- |
NAC transcription factor 066 [Jatropha curcas] |
| 9 |
Hb_001675_030 |
0.245799222 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_004899_310 |
0.2472711187 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_012807_130 |
0.2534096582 |
- |
- |
PREDICTED: lactoylglutathione lyase [Jatropha curcas] |
| 12 |
Hb_172632_050 |
0.2539412519 |
- |
- |
PREDICTED: uncharacterized protein LOC105646134 [Jatropha curcas] |
| 13 |
Hb_001999_240 |
0.2607662009 |
- |
- |
PREDICTED: josephin-like protein [Populus euphratica] |
| 14 |
Hb_002249_070 |
0.2608353293 |
- |
- |
gcn4-complementing protein, putative [Ricinus communis] |
| 15 |
Hb_000856_050 |
0.2617144886 |
- |
- |
PREDICTED: HIPL1 protein-like [Jatropha curcas] |
| 16 |
Hb_000450_160 |
0.2623107862 |
- |
- |
hypothetical protein JCGZ_16746 [Jatropha curcas] |
| 17 |
Hb_006922_090 |
0.2630850283 |
- |
- |
PREDICTED: interaptin [Jatropha curcas] |
| 18 |
Hb_000776_060 |
0.2638622118 |
- |
- |
histone H4 [Zea mays] |
| 19 |
Hb_002035_110 |
0.2638986291 |
transcription factor |
TF Family: TRAF |
Regulatory protein NPR1, putative [Ricinus communis] |
| 20 |
Hb_005663_070 |
0.2642813001 |
- |
- |
dehydrin protein [Manihot esculenta] |