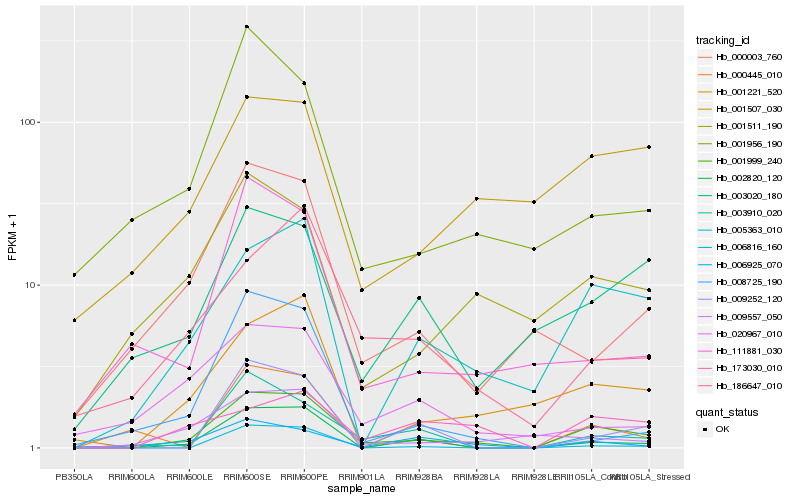
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001999_240 |
0.0 |
- |
- |
PREDICTED: josephin-like protein [Populus euphratica] |
| 2 |
Hb_002820_120 |
0.2086393622 |
transcription factor |
TF Family: ERF |
AP2 domain-containing transcription factor family protein [Populus trichocarpa] |
| 3 |
Hb_005363_010 |
0.2089646758 |
- |
- |
hypothetical protein JCGZ_13835 [Jatropha curcas] |
| 4 |
Hb_001221_520 |
0.2261879592 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_003910_020 |
0.2431964449 |
- |
- |
PREDICTED: reticulon-like protein B13 [Jatropha curcas] |
| 6 |
Hb_009557_050 |
0.2467030226 |
transcription factor |
TF Family: C2H2 |
zinc finger family protein [Populus trichocarpa] |
| 7 |
Hb_186647_010 |
0.2509676105 |
- |
- |
- |
| 8 |
Hb_000003_760 |
0.254922972 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA26-like [Jatropha curcas] |
| 9 |
Hb_006925_070 |
0.2556392287 |
- |
- |
hypothetical protein JCGZ_20652 [Jatropha curcas] |
| 10 |
Hb_001511_190 |
0.2559856161 |
- |
- |
PREDICTED: probable protein phosphatase 2C 25 [Jatropha curcas] |
| 11 |
Hb_006816_160 |
0.2604508837 |
- |
- |
hypothetical protein RCOM_1434920 [Ricinus communis] |
| 12 |
Hb_173030_010 |
0.2607662009 |
- |
- |
hypothetical protein RCOM_1321060 [Ricinus communis] |
| 13 |
Hb_003020_180 |
0.2633581925 |
- |
- |
PREDICTED: probable protein phosphatase 2C 24 [Jatropha curcas] |
| 14 |
Hb_111881_030 |
0.2634878198 |
- |
- |
PREDICTED: uncharacterized protein LOC105645255 [Jatropha curcas] |
| 15 |
Hb_009252_120 |
0.2660780367 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001956_190 |
0.267788563 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1 [Jatropha curcas] |
| 17 |
Hb_001507_030 |
0.2714019563 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 2 [Jatropha curcas] |
| 18 |
Hb_008725_190 |
0.2764083689 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein 5-like [Jatropha curcas] |
| 19 |
Hb_020967_010 |
0.2799388548 |
- |
- |
- |
| 20 |
Hb_000445_010 |
0.283239146 |
- |
- |
PREDICTED: uncharacterized protein LOC105113411 [Populus euphratica] |