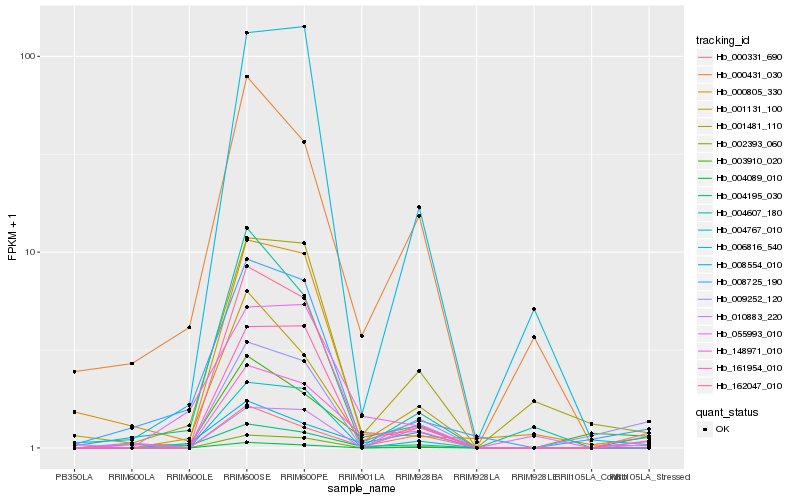
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_148971_010 |
0.0 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 2 |
Hb_162047_010 |
0.1879039676 |
- |
- |
PREDICTED: protein indeterminate-domain 16 [Jatropha curcas] |
| 3 |
Hb_003910_020 |
0.1994742931 |
- |
- |
PREDICTED: reticulon-like protein B13 [Jatropha curcas] |
| 4 |
Hb_004195_030 |
0.2175160443 |
- |
- |
hypothetical protein POPTR_0006s17880g, partial [Populus trichocarpa] |
| 5 |
Hb_000331_690 |
0.2196648668 |
- |
- |
PREDICTED: F-box/WD repeat-containing protein 11-like [Jatropha curcas] |
| 6 |
Hb_002393_060 |
0.2270648742 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor WER-like [Jatropha curcas] |
| 7 |
Hb_004089_010 |
0.2296735229 |
- |
- |
hypothetical protein MIMGU_mgv1a018902mg [Erythranthe guttata] |
| 8 |
Hb_008725_190 |
0.230062796 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein 5-like [Jatropha curcas] |
| 9 |
Hb_000431_030 |
0.2305453129 |
transcription factor |
TF Family: C2H2 |
zinc finger family protein [Populus trichocarpa] |
| 10 |
Hb_001131_100 |
0.2366957305 |
- |
- |
PREDICTED: uncharacterized protein LOC105638386 [Jatropha curcas] |
| 11 |
Hb_004767_010 |
0.2370164577 |
- |
- |
hypothetical protein CICLE_v10010736mg [Citrus clementina] |
| 12 |
Hb_001481_110 |
0.2405329301 |
- |
- |
S-adenosylmethionine synthetase 1 family protein [Populus trichocarpa] |
| 13 |
Hb_009252_120 |
0.2406680277 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_010883_220 |
0.2420170891 |
- |
- |
PREDICTED: flavonol synthase/flavanone 3-hydroxylase-like [Prunus mume] |
| 15 |
Hb_161954_010 |
0.2427178055 |
- |
- |
hypothetical protein JCGZ_05404 [Jatropha curcas] |
| 16 |
Hb_004607_180 |
0.2447753826 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_006816_540 |
0.2458346907 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET16-like [Jatropha curcas] |
| 18 |
Hb_008554_010 |
0.2469143701 |
- |
- |
- |
| 19 |
Hb_000805_330 |
0.2473105174 |
- |
- |
PREDICTED: ricin-like [Elaeis guineensis] |
| 20 |
Hb_055993_010 |
0.2473793912 |
- |
- |
PREDICTED: uncharacterized protein LOC104883809 [Beta vulgaris subsp. vulgaris] |