| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
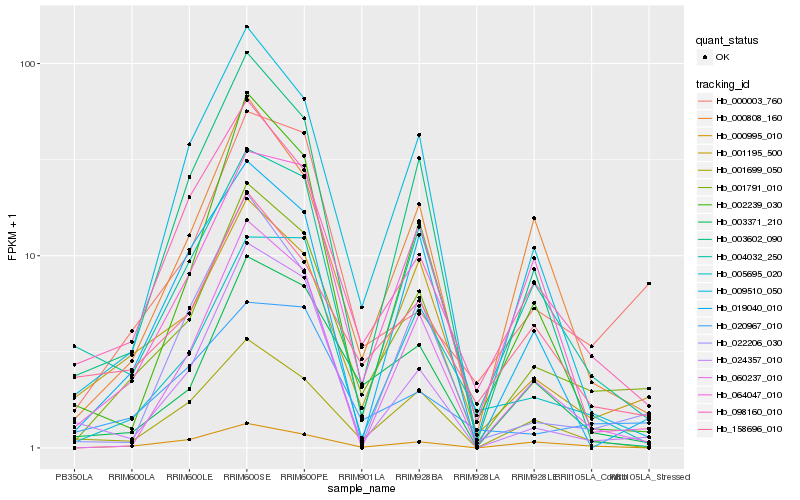
Hb_001791_010 |
0.0 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g63710 [Jatropha curcas] |
| 2 |
Hb_001195_500 |
0.1389117587 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein indeterminate-domain 2-like [Jatropha curcas] |
| 3 |
Hb_003371_210 |
0.1516159581 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 4 |
Hb_098160_010 |
0.1548670624 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X2 [Jatropha curcas] |
| 5 |
Hb_009510_050 |
0.1617682633 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000808_160 |
0.1631568616 |
- |
- |
PREDICTED: uncharacterized protein LOC105647443 [Jatropha curcas] |
| 7 |
Hb_004032_250 |
0.1671601204 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000003_760 |
0.1680716912 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA26-like [Jatropha curcas] |
| 9 |
Hb_158696_010 |
0.170502433 |
- |
- |
PREDICTED: putative disease resistance protein RGA4, partial [Jatropha curcas] |
| 10 |
Hb_001699_050 |
0.1721994834 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0005s11410g [Populus trichocarpa] |
| 11 |
Hb_003602_090 |
0.1724550517 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA9-like [Jatropha curcas] |
| 12 |
Hb_060237_010 |
0.1727071932 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002239_030 |
0.1733718615 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 4 [Jatropha curcas] |
| 14 |
Hb_000995_010 |
0.1738517415 |
- |
- |
PREDICTED: uncharacterized protein LOC105635846 [Jatropha curcas] |
| 15 |
Hb_022206_030 |
0.17881627 |
- |
- |
Auxin-induced in root cultures protein 12 precursor, putative [Ricinus communis] |
| 16 |
Hb_019040_010 |
0.179621633 |
- |
- |
PREDICTED: indole-3-acetic acid-induced protein ARG7 [Jatropha curcas] |
| 17 |
Hb_020967_010 |
0.1803080961 |
- |
- |
- |
| 18 |
Hb_005695_020 |
0.1803360581 |
- |
- |
PREDICTED: low affinity sulfate transporter 3 [Jatropha curcas] |
| 19 |
Hb_024357_010 |
0.1806543071 |
- |
- |
PREDICTED: putative disease resistance protein RGA4, partial [Jatropha curcas] |
| 20 |
Hb_064047_010 |
0.180702674 |
- |
- |
conserved hypothetical protein [Ricinus communis] |