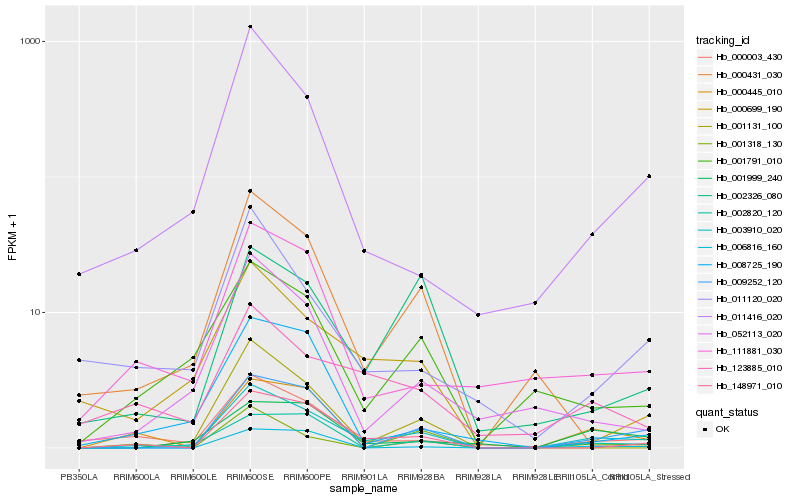
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003910_020 |
0.0 |
- |
- |
PREDICTED: reticulon-like protein B13 [Jatropha curcas] |
| 2 |
Hb_009252_120 |
0.1993272835 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_148971_010 |
0.1994742931 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 4 |
Hb_000003_430 |
0.2140383395 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 5 |
Hb_002820_120 |
0.225462687 |
transcription factor |
TF Family: ERF |
AP2 domain-containing transcription factor family protein [Populus trichocarpa] |
| 6 |
Hb_011416_020 |
0.2307840599 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001791_010 |
0.2320091391 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g63710 [Jatropha curcas] |
| 8 |
Hb_052113_020 |
0.2368511794 |
- |
- |
hypothetical protein CISIN_1g022501mg [Citrus sinensis] |
| 9 |
Hb_111881_030 |
0.2371282644 |
- |
- |
PREDICTED: uncharacterized protein LOC105645255 [Jatropha curcas] |
| 10 |
Hb_000445_010 |
0.2424581036 |
- |
- |
PREDICTED: uncharacterized protein LOC105113411 [Populus euphratica] |
| 11 |
Hb_002326_080 |
0.2428701294 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF1.6 [Jatropha curcas] |
| 12 |
Hb_001999_240 |
0.2431964449 |
- |
- |
PREDICTED: josephin-like protein [Populus euphratica] |
| 13 |
Hb_000699_190 |
0.2432242523 |
- |
- |
unknown [Populus trichocarpa] |
| 14 |
Hb_123885_010 |
0.2446990984 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 15 |
Hb_006816_160 |
0.2463496641 |
- |
- |
hypothetical protein RCOM_1434920 [Ricinus communis] |
| 16 |
Hb_001318_130 |
0.2487261941 |
- |
- |
PREDICTED: short-chain dehydrogenase reductase 3b-like [Populus euphratica] |
| 17 |
Hb_001131_100 |
0.2502189073 |
- |
- |
PREDICTED: uncharacterized protein LOC105638386 [Jatropha curcas] |
| 18 |
Hb_011120_020 |
0.2515918757 |
- |
- |
PREDICTED: casein kinase I-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_000431_030 |
0.2519803735 |
transcription factor |
TF Family: C2H2 |
zinc finger family protein [Populus trichocarpa] |
| 20 |
Hb_008725_190 |
0.2530351638 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein 5-like [Jatropha curcas] |