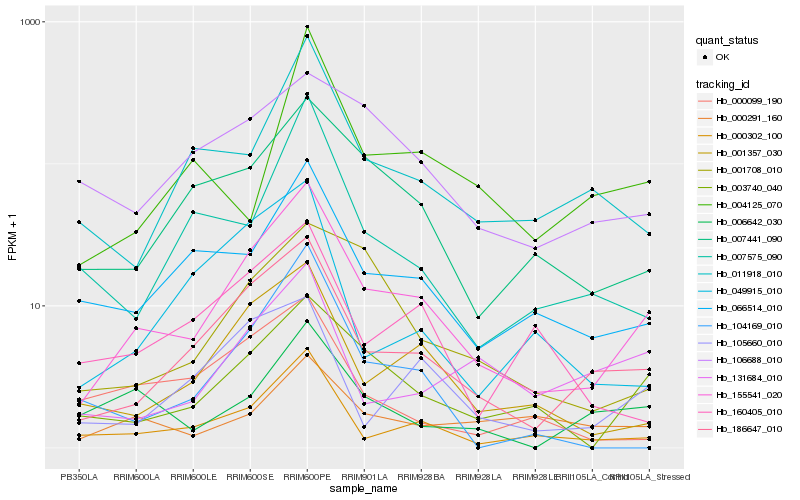
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_155541_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_186647_010 |
0.1410604916 |
- |
- |
- |
| 3 |
Hb_003740_040 |
0.1840585583 |
- |
- |
Ubiquitin carboxyl-terminal hydrolase isozyme L3, putative [Ricinus communis] |
| 4 |
Hb_001357_030 |
0.1851371781 |
- |
- |
hypothetical protein F383_04880 [Gossypium arboreum] |
| 5 |
Hb_000302_100 |
0.1863318406 |
- |
- |
PREDICTED: uncharacterized protein LOC105766209 [Gossypium raimondii] |
| 6 |
Hb_007441_090 |
0.2009522241 |
- |
- |
nucleoside diphosphate kinase 1 [Hevea brasiliensis] |
| 7 |
Hb_066514_010 |
0.2049041157 |
- |
- |
hypothetical protein RCOM_0204720 [Ricinus communis] |
| 8 |
Hb_011918_010 |
0.221278721 |
- |
- |
hypothetical protein glysoja_041948 [Glycine soja] |
| 9 |
Hb_105660_010 |
0.2226611467 |
- |
- |
PREDICTED: potassium channel AKT6-like [Jatropha curcas] |
| 10 |
Hb_001708_010 |
0.2331168653 |
- |
- |
PREDICTED: putative disease resistance protein At4g11170 [Malus domestica] |
| 11 |
Hb_006642_030 |
0.2374515477 |
- |
- |
PREDICTED: germin-like protein subfamily 3 member 2 [Jatropha curcas] |
| 12 |
Hb_007575_090 |
0.2418318408 |
- |
- |
putative. proteolipid subunit of vacuolar H+ ATPase, partial [Zea mays] |
| 13 |
Hb_049915_010 |
0.242416237 |
- |
- |
PREDICTED: uncharacterized serine-rich protein C215.13 [Jatropha curcas] |
| 14 |
Hb_131684_010 |
0.2427154954 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_160405_010 |
0.2427562218 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 16 |
Hb_000291_160 |
0.2441404309 |
- |
- |
PREDICTED: uncharacterized protein LOC105638863 [Jatropha curcas] |
| 17 |
Hb_004125_070 |
0.2454935428 |
- |
- |
Glycolipid transfer protein, putative [Ricinus communis] |
| 18 |
Hb_106688_010 |
0.2462324809 |
- |
- |
aspartyl-tRNA synthetase, putative [Ricinus communis] |
| 19 |
Hb_104169_010 |
0.2476449131 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 20 |
Hb_000099_190 |
0.2501529001 |
- |
- |
PREDICTED: putative cell division cycle ATPase [Jatropha curcas] |