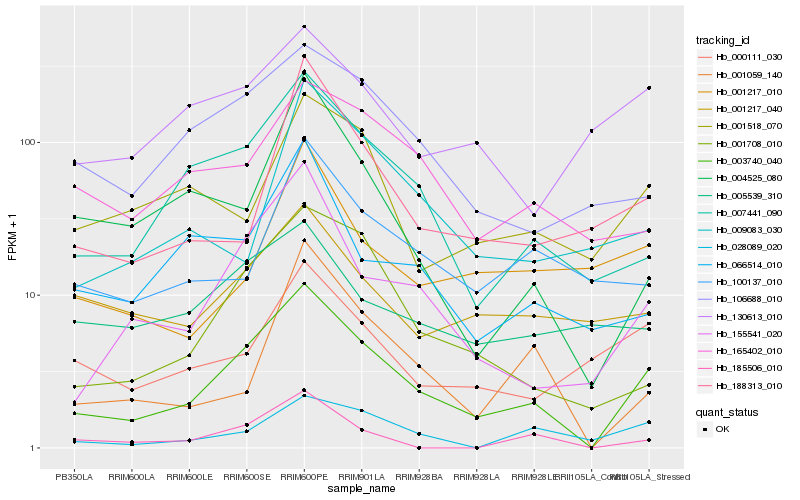
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003740_040 |
0.0 |
- |
- |
Ubiquitin carboxyl-terminal hydrolase isozyme L3, putative [Ricinus communis] |
| 2 |
Hb_155541_020 |
0.1840585583 |
- |
- |
- |
| 3 |
Hb_007441_090 |
0.1898469253 |
- |
- |
nucleoside diphosphate kinase 1 [Hevea brasiliensis] |
| 4 |
Hb_001708_010 |
0.1975641634 |
- |
- |
PREDICTED: putative disease resistance protein At4g11170 [Malus domestica] |
| 5 |
Hb_106688_010 |
0.2158385407 |
- |
- |
aspartyl-tRNA synthetase, putative [Ricinus communis] |
| 6 |
Hb_001059_140 |
0.2297017364 |
- |
- |
hypothetical protein POPTR_0004s03390g [Populus trichocarpa] |
| 7 |
Hb_066514_010 |
0.2319394425 |
- |
- |
hypothetical protein RCOM_0204720 [Ricinus communis] |
| 8 |
Hb_028089_020 |
0.2353057418 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Prunus mume] |
| 9 |
Hb_165402_010 |
0.2353911531 |
- |
- |
PREDICTED: cullin-1 isoform X3 [Nicotiana sylvestris] |
| 10 |
Hb_188313_010 |
0.2355067775 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: carboxymethylenebutenolidase homolog [Jatropha curcas] |
| 11 |
Hb_000111_030 |
0.236685545 |
- |
- |
PREDICTED: uncharacterized protein LOC104094030 [Nicotiana tomentosiformis] |
| 12 |
Hb_185506_010 |
0.2369698797 |
- |
- |
choline monooxygenase, putative [Ricinus communis] |
| 13 |
Hb_001518_070 |
0.2375150223 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_100137_010 |
0.2387631447 |
- |
- |
hypothetical protein JCGZ_17848 [Jatropha curcas] |
| 15 |
Hb_004525_080 |
0.2393357514 |
- |
- |
Sugar transporter ERD6-like 6 [Morus notabilis] |
| 16 |
Hb_001217_040 |
0.2416543593 |
- |
- |
hypothetical protein MIMGU_mgv1a0002421mg, partial [Erythranthe guttata] |
| 17 |
Hb_009083_030 |
0.2448814294 |
- |
- |
hypothetical protein POPTR_0007s14760g [Populus trichocarpa] |
| 18 |
Hb_130613_010 |
0.2461645708 |
- |
- |
Charged multivesicular body protein 2a, putative [Ricinus communis] |
| 19 |
Hb_005539_310 |
0.2480242159 |
- |
- |
basic helix-loop-helix-containing protein, putative [Ricinus communis] |
| 20 |
Hb_001217_010 |
0.2495787265 |
- |
- |
hypothetical protein JCGZ_04022 [Jatropha curcas] |