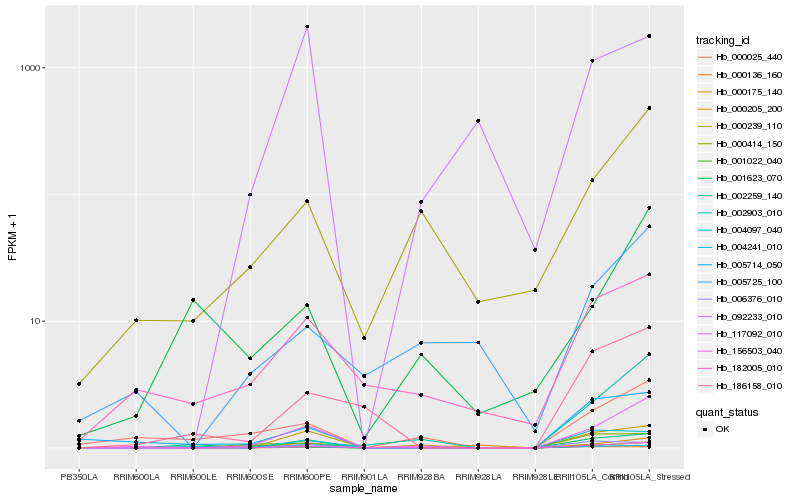
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_117092_010 |
0.0 |
- |
- |
acyl-CoA oxidase, putative [Ricinus communis] |
| 2 |
Hb_000205_200 |
0.2034363719 |
- |
- |
- |
| 3 |
Hb_000239_110 |
0.259146845 |
- |
- |
PREDICTED: adenine phosphoribosyltransferase 1-like [Jatropha curcas] |
| 4 |
Hb_000025_440 |
0.2705181165 |
- |
- |
PREDICTED: ankyrin repeat-containing protein P16F5.05c-like [Populus euphratica] |
| 5 |
Hb_000175_140 |
0.2787364931 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_186158_010 |
0.2816232812 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_002259_140 |
0.2818755381 |
- |
- |
- |
| 8 |
Hb_004097_040 |
0.2850154357 |
- |
- |
PREDICTED: aspartate--tRNA ligase, mitochondrial [Jatropha curcas] |
| 9 |
Hb_000414_150 |
0.2852358619 |
- |
- |
mta/sah nucleosidase, putative [Ricinus communis] |
| 10 |
Hb_006376_010 |
0.2916336102 |
- |
- |
PREDICTED: uncharacterized protein LOC104090718 [Nicotiana tomentosiformis] |
| 11 |
Hb_005725_100 |
0.293693172 |
- |
- |
hypothetical protein 30 [Hevea brasiliensis] |
| 12 |
Hb_000136_160 |
0.2937889785 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 13 |
Hb_182005_010 |
0.2952202355 |
- |
- |
PREDICTED: uncharacterized protein LOC105639166 [Jatropha curcas] |
| 14 |
Hb_004241_010 |
0.3021531517 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 15 |
Hb_005714_050 |
0.3023460873 |
transcription factor |
TF Family: B3 |
hypothetical protein POPTR_1461s00200g [Populus trichocarpa] |
| 16 |
Hb_092233_010 |
0.3031633596 |
- |
- |
RecName: Full=Profilin-3; AltName: Full=Pollen allergen Hev b 8.0201; AltName: Allergen=Hev b 8.0201 [Hevea brasiliensis] |
| 17 |
Hb_001022_040 |
0.3049900526 |
- |
- |
PREDICTED: uncharacterized protein LOC105634753 [Jatropha curcas] |
| 18 |
Hb_002903_010 |
0.3061547976 |
- |
- |
60S ribosomal protein L37-3 [Morus notabilis] |
| 19 |
Hb_156503_040 |
0.3064195735 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Jatropha curcas] |
| 20 |
Hb_001623_070 |
0.3073953542 |
- |
- |
cytochrome B5 isoform 1, putative [Ricinus communis] |