| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
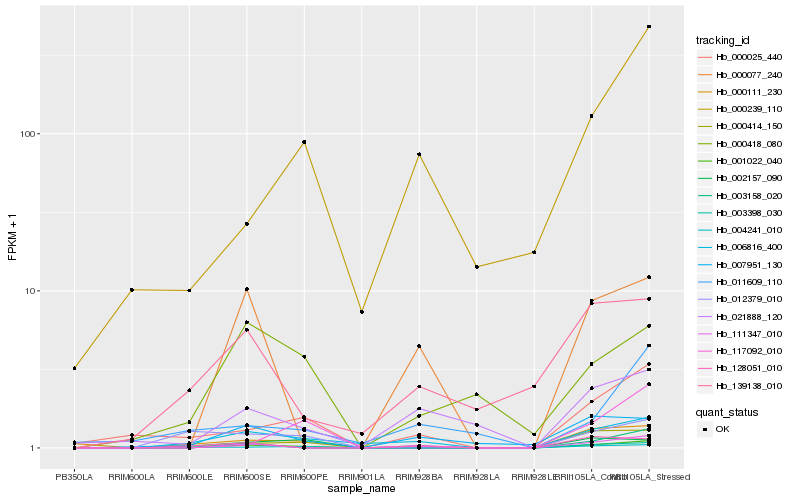
Hb_001022_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105634753 [Jatropha curcas] |
| 2 |
Hb_006816_400 |
0.1796359759 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 20 [Jatropha curcas] |
| 3 |
Hb_000025_440 |
0.2304412085 |
- |
- |
PREDICTED: ankyrin repeat-containing protein P16F5.05c-like [Populus euphratica] |
| 4 |
Hb_111347_010 |
0.2468356208 |
- |
- |
PREDICTED: uncharacterized protein LOC105781565 [Gossypium raimondii] |
| 5 |
Hb_004241_010 |
0.2523007286 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 6 |
Hb_000414_150 |
0.2569147115 |
- |
- |
mta/sah nucleosidase, putative [Ricinus communis] |
| 7 |
Hb_007951_130 |
0.2652522633 |
desease resistance |
Gene Name: UBN2_3 |
JHL25H03.6 [Jatropha curcas] |
| 8 |
Hb_021888_120 |
0.2800626981 |
- |
- |
PREDICTED: putative CCR4-associated factor 1 homolog 8 [Jatropha curcas] |
| 9 |
Hb_000111_230 |
0.2814180922 |
- |
- |
- |
| 10 |
Hb_000239_110 |
0.2841812691 |
- |
- |
PREDICTED: adenine phosphoribosyltransferase 1-like [Jatropha curcas] |
| 11 |
Hb_000418_080 |
0.2846873234 |
- |
- |
PREDICTED: uncharacterized protein LOC105124652 [Populus euphratica] |
| 12 |
Hb_000077_240 |
0.2875338491 |
- |
- |
hypothetical protein PHAVU_011G114500g, partial [Phaseolus vulgaris] |
| 13 |
Hb_003398_030 |
0.2972508219 |
- |
- |
PREDICTED: uncharacterized protein LOC104583892 [Brachypodium distachyon] |
| 14 |
Hb_002157_090 |
0.3003507635 |
- |
- |
hypothetical protein VITISV_041741 [Vitis vinifera] |
| 15 |
Hb_003158_020 |
0.3010504197 |
- |
- |
PREDICTED: uncharacterized protein LOC104647507 [Solanum lycopersicum] |
| 16 |
Hb_012379_010 |
0.3013722669 |
- |
- |
- |
| 17 |
Hb_117092_010 |
0.3049900526 |
- |
- |
acyl-CoA oxidase, putative [Ricinus communis] |
| 18 |
Hb_139138_010 |
0.3054793877 |
- |
- |
PREDICTED: uncharacterized protein LOC105650977 [Jatropha curcas] |
| 19 |
Hb_128051_010 |
0.3074385169 |
- |
- |
- |
| 20 |
Hb_011609_110 |
0.3132899661 |
- |
- |
PREDICTED: T-complex protein 1 subunit gamma-like [Jatropha curcas] |