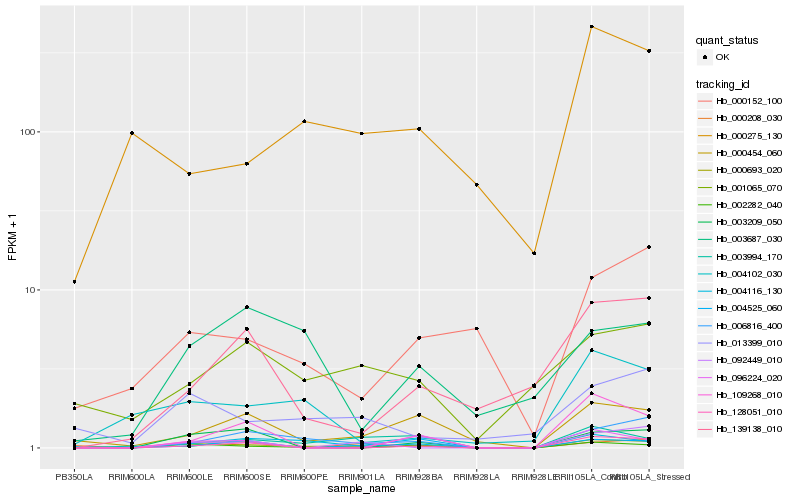
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000693_020 |
0.0 |
- |
- |
PREDICTED: VQ motif-containing protein 11 [Jatropha curcas] |
| 2 |
Hb_002282_040 |
0.2084990997 |
- |
- |
hypothetical protein VITISV_030841 [Vitis vinifera] |
| 3 |
Hb_006816_400 |
0.2242500717 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 20 [Jatropha curcas] |
| 4 |
Hb_004116_130 |
0.2360823436 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 5 |
Hb_000454_060 |
0.2589370005 |
- |
- |
Uncharacterized protein TCM_006778 [Theobroma cacao] |
| 6 |
Hb_128051_010 |
0.289149113 |
- |
- |
- |
| 7 |
Hb_013399_010 |
0.2947367311 |
- |
- |
PREDICTED: uncharacterized protein LOC105631404 isoform X2 [Jatropha curcas] |
| 8 |
Hb_003994_170 |
0.2980405156 |
- |
- |
Indole-3-acetic acid-induced protein ARG7, putative [Ricinus communis] |
| 9 |
Hb_003209_050 |
0.2995588513 |
- |
- |
hypothetical protein JCGZ_15683 [Jatropha curcas] |
| 10 |
Hb_096224_020 |
0.3028301808 |
- |
- |
hypothetical protein JCGZ_06980 [Jatropha curcas] |
| 11 |
Hb_004102_030 |
0.3064150733 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At5g63020 [Jatropha curcas] |
| 12 |
Hb_000208_030 |
0.308875565 |
- |
- |
- |
| 13 |
Hb_139138_010 |
0.3104914004 |
- |
- |
PREDICTED: uncharacterized protein LOC105650977 [Jatropha curcas] |
| 14 |
Hb_000152_100 |
0.3116014473 |
- |
- |
PREDICTED: probable protein phosphatase 2C 75 isoform X2 [Jatropha curcas] |
| 15 |
Hb_001065_070 |
0.3116874667 |
- |
- |
- |
| 16 |
Hb_004525_060 |
0.3137924339 |
- |
- |
- |
| 17 |
Hb_000275_130 |
0.3157527637 |
- |
- |
hypothetical protein CICLE_v10003066mg [Citrus clementina] |
| 18 |
Hb_109268_010 |
0.3184926688 |
- |
- |
PREDICTED: uncharacterized protein LOC105650977 [Jatropha curcas] |
| 19 |
Hb_003687_030 |
0.3224740808 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 20 |
Hb_092449_010 |
0.3224964258 |
- |
- |
- |